- Robert Crawford¹
- Hugo Sepulveda
- Xiang Li
- Leo J. Arteaga-Vazquez
- Isaac F. López-Moyado
- Melina Brunelli
- Lot Hernandez Espinosa
- Xiaojing Yue
- J. Carlos Angel
- Caitlin Brown
- Zhen Dong
- Natasha Jansz
- Fabio Puddu¹
- Aurélie Modat¹
- Jamie Scotcher¹
- Páidí Creed¹
- Patrick Kennedy
- Cindy Manriquez
- Samuel A Myers
- Geoffrey J. Faulkner
- Anjana Rao²
1 biomodal Ltd, The Trinity Building, Chesterford Research Park, Cambridge, UK;
2 Division of Signaling and Gene Expression, La Jolla Institute for Immunology, 9420 Athena Circle, La Jolla, CA, 34 92037, USA
We present a study investigating the effect of deletion of the epigenetic regulation enzyme O-GlcNAc transferase (OGT) on genome-wide 5mC and 5hmC levels in mouse embryonic stem cells (mESC) using a novel 6-base sequencing technology.
The O-GlcNAc transferase OGT interacts robustly with all three mammalian Ten-Eleven Translocation (TET) methylcytosine dioxygenases. Using duet evoC 6-base sequencing (enabling individual discrimination of A, C, T, G, 5mC and 5hmC), we show that deletion of the Ogt gene in mESC results in a widespread increase in the TET product 5hmC in both euchromatic and heterochromatic compartments, with concomitant reduction of the TET substrate 5mC at the same genomic regions. mESC treated with an OGT inhibitor also displayed increased 5hmC demonstrating OGT enzymatic activity is needed to suppress TET activity. This indicates that OGT restrains TET activity and limits untoward DNA demethylation in a manner that requires the TET-OGT interaction and the catalytic activity of OGT. DNA hypomethylation in OGT-deficient cells was accompanied by de-repression of transposable elements (TEs) predominantly located in heterochromatin.
We suggest that OGT protects the genome against TET-mediated DNA demethylation and loss of heterochromatin integrity, preventing the aberrant increase in TE expression noted in cancer, autoimmune-inflammatory diseases, cellular senescence and ageing.
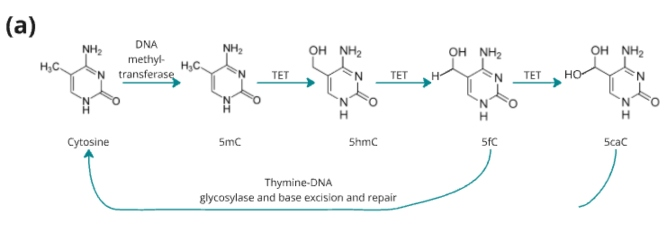
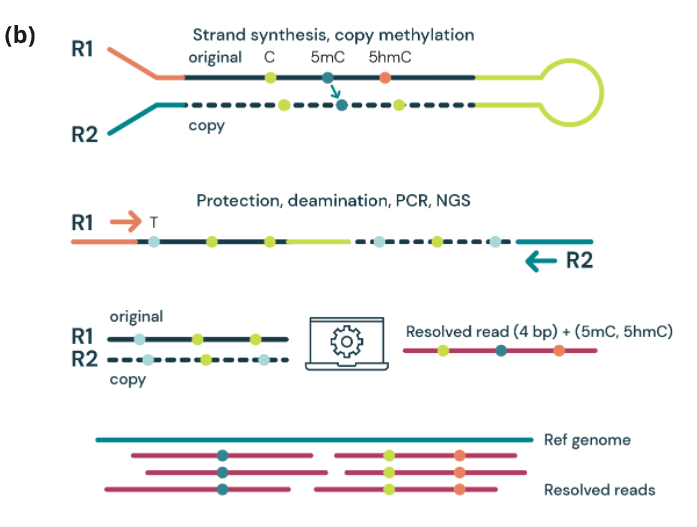
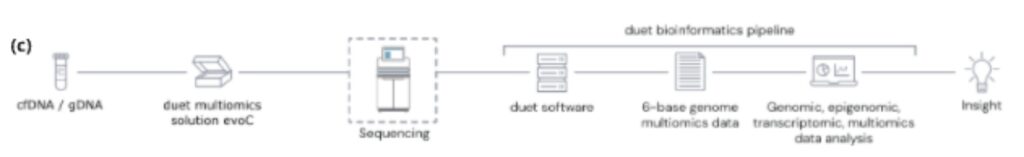
Figure 1. (a) TET-mediated demethylation pathway
(b) duet multiomics solution evoC – a 6-base sequencing technology that reads all four canonical bases plus 5mC and 5hmC¹ via strand copy, 5mC copy and 5mC + 5hmC protection enzymatic steps.
(c) The duet multiomics solution evoC works as an end-to-end solution comprising reagents & bioinformatics pipeline
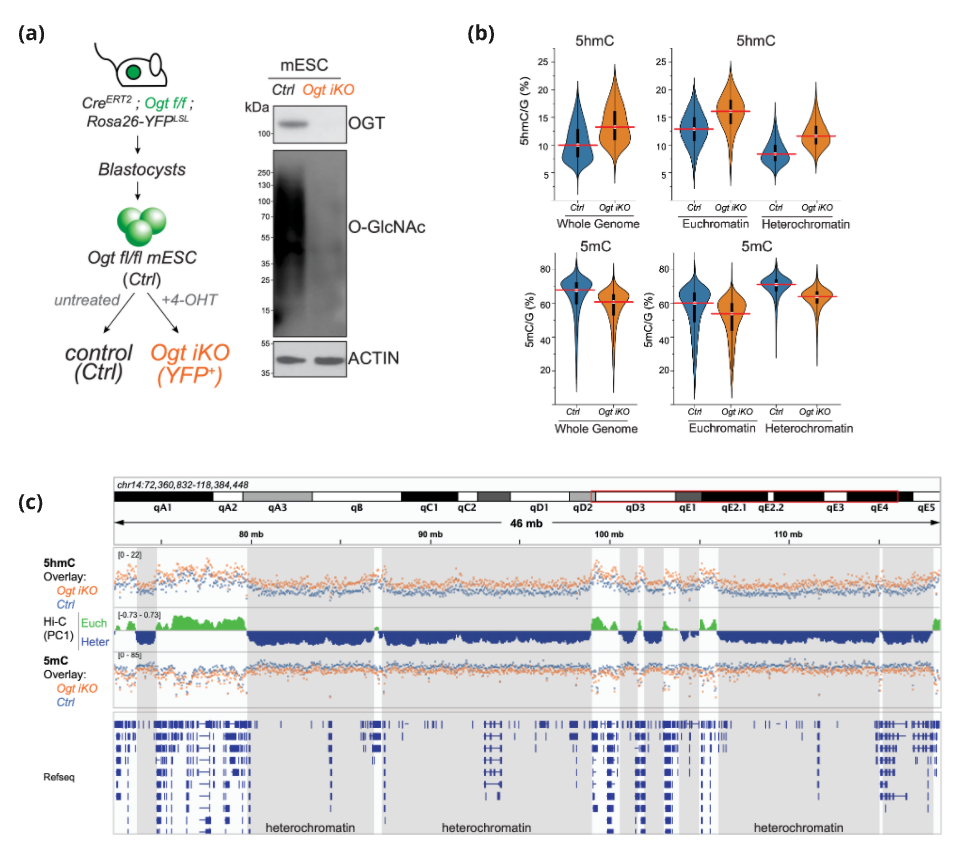
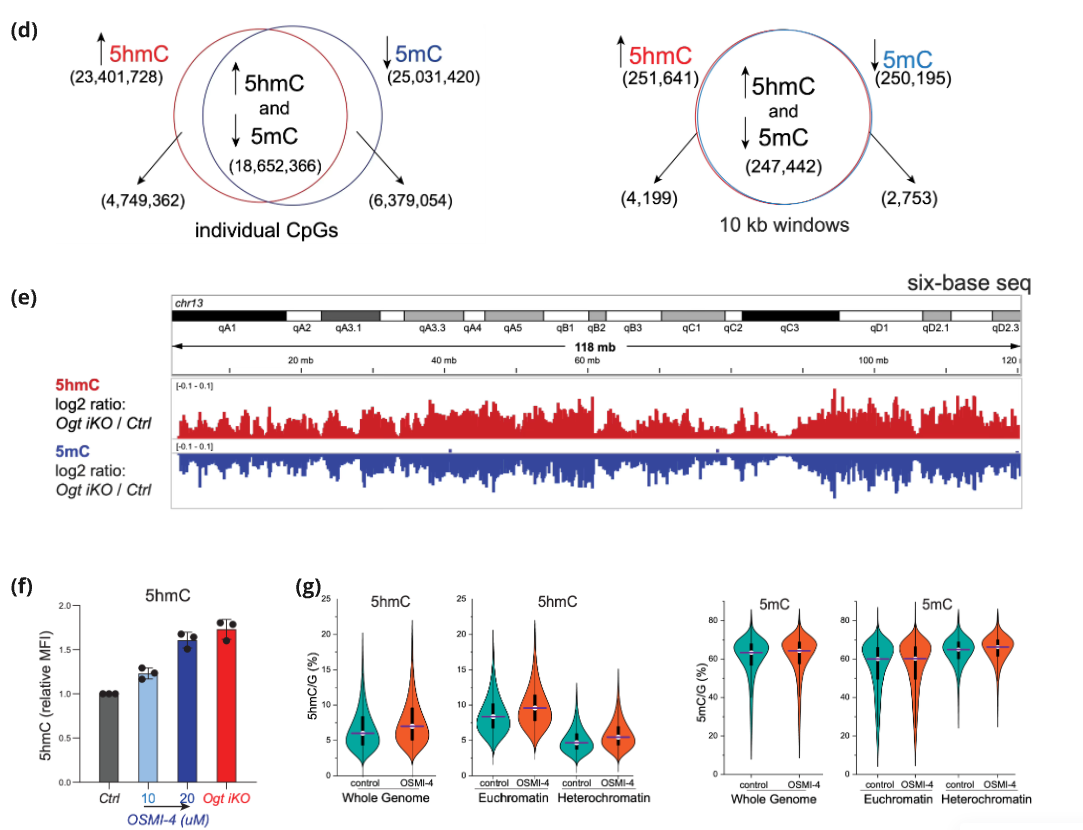
Figure 2. (a) Left. Generation of Ogt iKO mESC. Right. Western blots showing loss of OGT protein (top) and O-GlcNAc modification (bot) in whole-cell lysates.
(b) 6-base sequencing shows increased 5hmC (top) and decreased 5mC (bot) in Ogt iKO compared to Ogt fl (Ctrl) mESC at whole genome resolution (left) or chromatin compartments (right).
(c) Genome browser view of 5hmC (top) and 5mC (bottom) in overlaid tracks of Ogt iKO (orange) and Ctrl Ogt fl (blue) mESC. 10 kb windows averaged. Euchromatin (green), heterochromatin (blue).
(d) Venn diagrams showing overlap between gaining 5hmC (red) and losing 5mC (blue) in Ogt iKO mESC vs Ctrl Ogt fl mESC for individual CpGs left or 10kb windows right.
(e) Genome browser view showing reciprocal gain of 5hmC (red) and loss of 5mC (blue) in Ogt iKO vs Ctrl Ogt fl mESC. Plotted: log2 ratios (Ogt iKO/ Ctrl Ogt fl) for 5hmC (top) and 5mC (bottom).
(f) Increase of 5hmC after OGT inhibition with OSMI-4. 5hmC measured by flow cytometry after 4 days of OSMI-4 treatment. Bar graphs represent the mean ± s.d. from three independent experiments.
(g) Violin plots showing increased 5hmC levels (left) but unaltered 5mC levels (right – likely due to earlier earlier growth arrest) in OSMI-4-treated versus control mES cells.
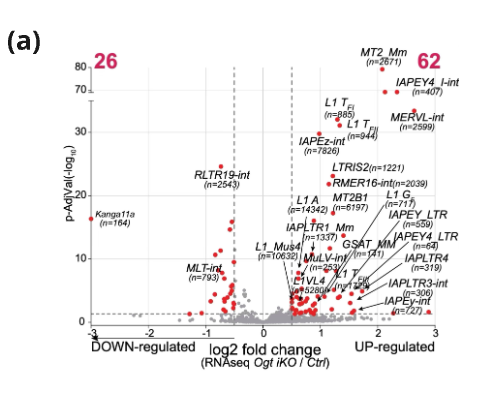
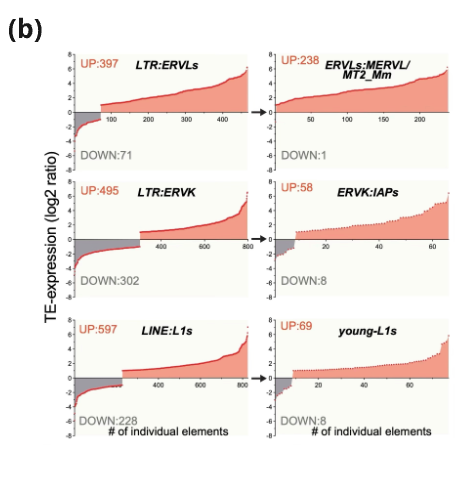
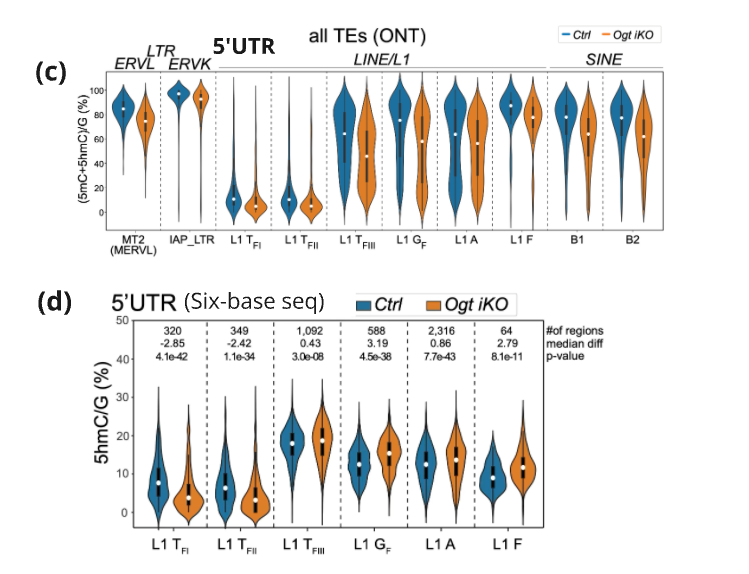
Figure 3. (a) Volcano plot of RNA-seq data showing differential TE expression in Ogt iKO vs Ctrl Ogt fl mESC. Red dots: transcripts exhibiting ≥ twofold expression change.
(b) RNA-seq data of Ogt iKO vs Ctrl Ogt fl mES cells, analysed for differential expression of uniquely mapped TEs. No. of upregulated (red) & downregulated (gray) TEs.
(c) modC (5mC+5hmC) levels of TE subfamilies in Ogt fl (blue) vs Ogt iKO (orange) mES cells (ONT)
(d) 5hmC levels at 5′ UTRs of indicated L1 families in Ctrl Ogt fl and Ogt iKO mES cells; difference of median 5hmC levels between conditions (Ogt iKO − Ctrl) displayed on top
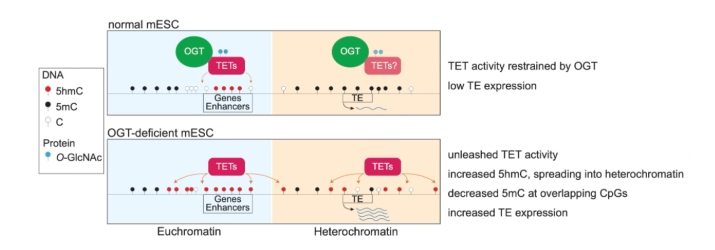
We report here that OGT globally restrains TET enzymatic activity and maintains DNA methylation genome-wide, in a manner that depends on OGT catalytic activity and the TET–OGT interaction (illustrated below). TET activity was unleashed upon Ogt gene deletion or OGT inhibition, resulting in ongoing demethylation defined by parallel increases in 5hmC and decreases in 5mC at overlapping CpGs, observed using duet multiomics solution evoC 6-base sequencing. The discovery (in press²) reveals a novel mechanism for maintaining genomic stability, with important implications for both development and disease.
- Füllgrabe J. et al. Simultaneous sequencing of genetic and epigenetic bases in DNA. Nat Biotechnol. 2023 Oct;41(10):1457-1464.
- Sepulveda, H. et al. OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide. Nat. Struct. Mol. Biol. (2025 – in press)
Figure 1. duet multiomics solution evoC is a 6-base calling technology that reads all four canonical bases plus 5mC and 5hmC.




