Resources
Resource types
- Blog post
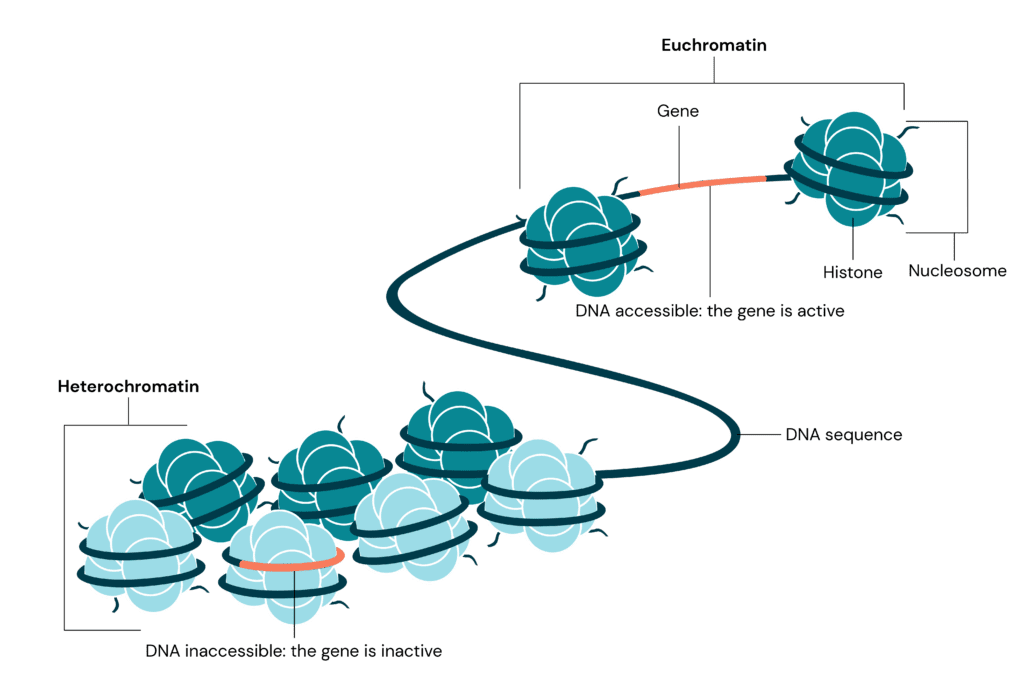
Heterochromatin is a condensed form of chromatin that plays an essential role in maintaining genomic stability. Disruptions in its formation can lead to disease. We explore the mysteries of heterochromatin and its vital role in genome organisation.
- News article
biomodal has just announced it has filed a lawsuit against New England Biolabs in the United States District Court for the District of Massachusetts.
- Technical note
DNA contains molecular information encoded in both genetic and epigenetic bases, essential for understanding biology.
- Poster
Liquid biopsy for profiling of cell free DNA (cfDNA) in blood holds huge promise to transform how we experience and manage cancer by early detection and identification of residual disease and subtype.
- Poster
Formalin-fixed, paraffin-embedded (FFPE) specimens are commonly used for long-term storage in research and clinical settings, including immunohistochemistry, oncology, and genomics.
- Poster
To address the difficulties of analysing methylation data, we present modality, an efficient and scalable analysis package for 5- and 6-base genomes.
- Poster
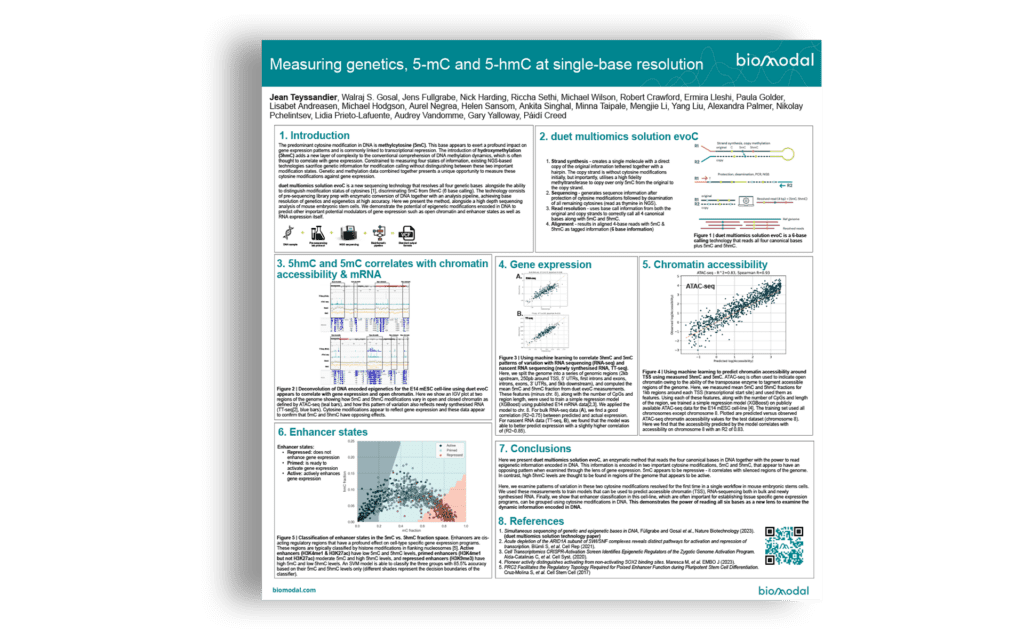
Using the 6‑base genome, genetics, 5mC and 5hmC, to gain multimodal insights into genome organisation and gene regulation
- Blog post
An Epigenome Wide Association Study (EWAS) investigates the relationship between epigenetic modifications and traits/diseases. Learn why they are important.
- Webinar
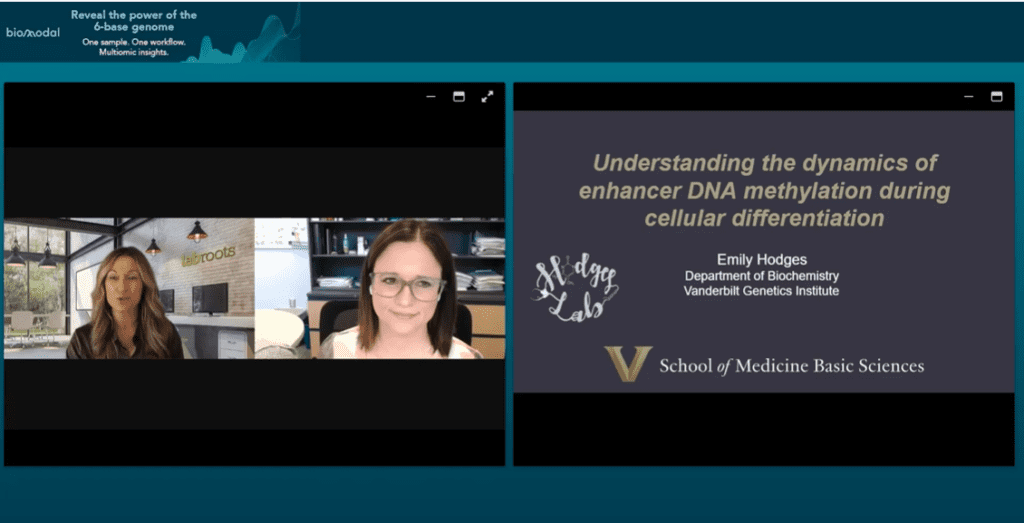
Join Dr Emily Hodges, assistant professor of biochemistry at Vanderbilt University, to learn more about how the Hodges Lab utilises 6-base genome to investigate the dynamics of enhancer DNA methylation during cellular differentiation.
- Blog post
Formalin-Fixed Paraffin-Embedded (FFPE) tissue plays a critical role in histopathological diagnosis, and it is particularly valuable in the diagnosis of diseases like cancer.
- Technical note
FFPE cancer samples are an incredibly important and clinically relevant information source for directly studying the causes and consequences of the disease. This technical note shows how duet evoC can identify DNA methylation markers from FFPE samples.
- Poster
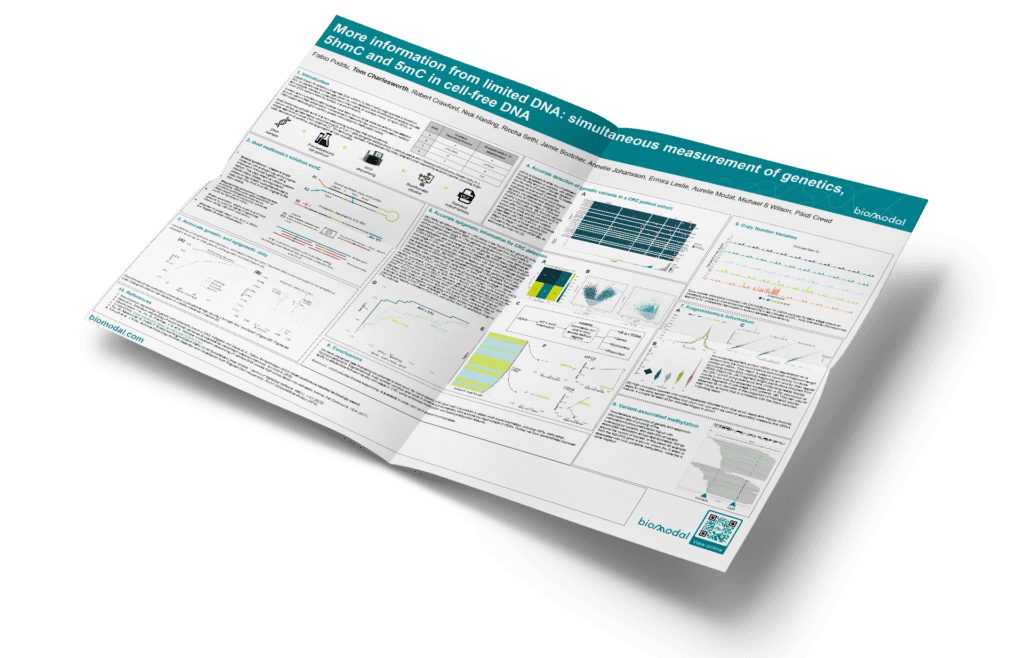
Liquid biopsy for profiling of cell free DNA (cfDNA) in blood holds huge promise to transform how we experience and manage. But can you get More information from limited DNA: simultaneous measurement of genetics, 5hmC and 5mC in cell-free DNA?