- Aurelie Modat
- Robert Crawford
- Fabio Puddu
- Annelie Johansson
- Riccha Sethi
- Tim Beech
- Tom Charlesworth
- Páidí Creed
Formalin-fixed, paraffin-embedded (FFPE) specimens are commonly used for long-term storage in research and clinical settings, including immunohistochemistry, oncology, and genomics. These samples have the potential to add value to studies incorporating both genomic and epigenomic information such as cytosine modifications (5mC & 5hmC), providing deeper insights into gene activation and silencing pathways and enabling a better understanding of cancer mechanisms. This combined data from FFPE samples enhances biological insights. However, formalin fixation can cause DNA damage, such as deamination, fragmentation and crosslinking, reducing data quality in next-generation sequencing (NGS).
Here we apply a novel 6-base sequencing technology (duet multiomics solution evoC) to DNA extracted from both controlled formalin-damaged standards and FFPE samples from colorectal and lung cancers. This method simultaneously detects cytosine modifications (5mC, 5hmC) and canonical bases (A, C, G, T) in a single workflow, reducing information loss and preserving C-to-T transition detection. This study evaluates the quality of 6-base genomes from those samples and the impact of formalin exposure on epigenetic sequencing results, comparing FFPE and fresh-frozen cancer samples to assess fixation-induced changes in DNA methylation profiles. Additionally, we look at how to achieve sufficient yield for formalin-damaged samples by increasing the number of PCR cycles.
- Strand synthesis: creates a single molecule with a direct copy of the original information tethered together with a hairpin. The copy strand is without cytosine modifications initially, but importantly, utilises a high fidelity methyltransferase to copy over only 5mC from the original to the copy strand.
- Sequencing – generates sequence information after protection of cytosine modifications followed by deamination of all remaining cytosines (read as thymine in NGS).
- Read resolution – uses base call information from both the original and copy strands to correctly call all 4 canonical bases along with 5mC and 5hmC.
- Alignment – results in aligned 4-base reads with 5mC & 5hmC as tagged information (6 base information)
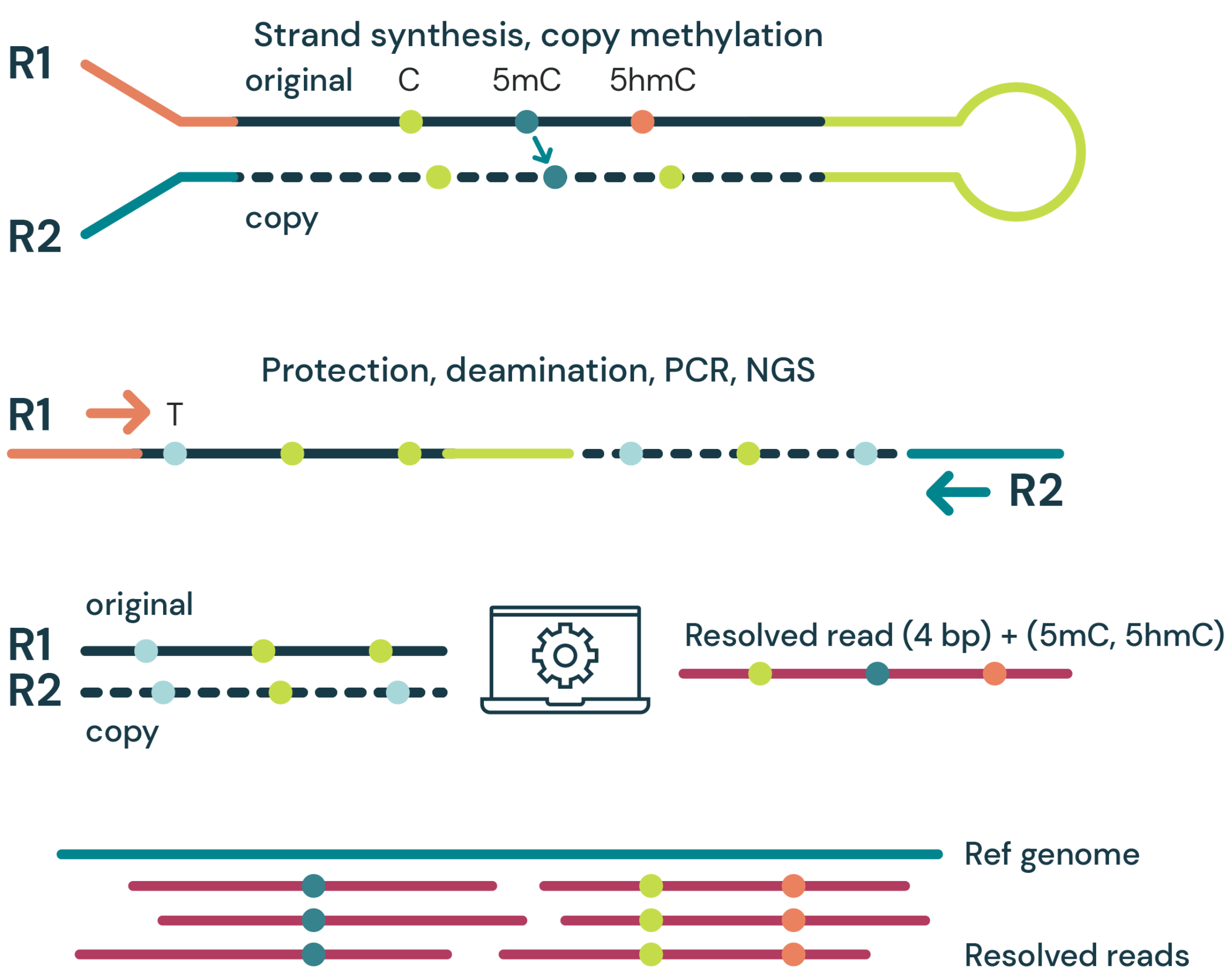
duet multiomics solution evoC is a 6-base calling technology that reads all four canonical bases plus 5mC and 5hmC.
duet multomics solution evoC applied to formalin-damaged DNA. Libraries were generated in triplicate with 10ng (minimum input) and 80ng (maximum input) of Horizon Quantitative Multiplex Reference Standard (QMRS) DNA either untreated or formalin-compromised DNA (fcDNA) with mild, moderate and severe damage. 4 libraries were also generated from matched FFPE and fresh frozen (FF) cancer patients at 80ng.
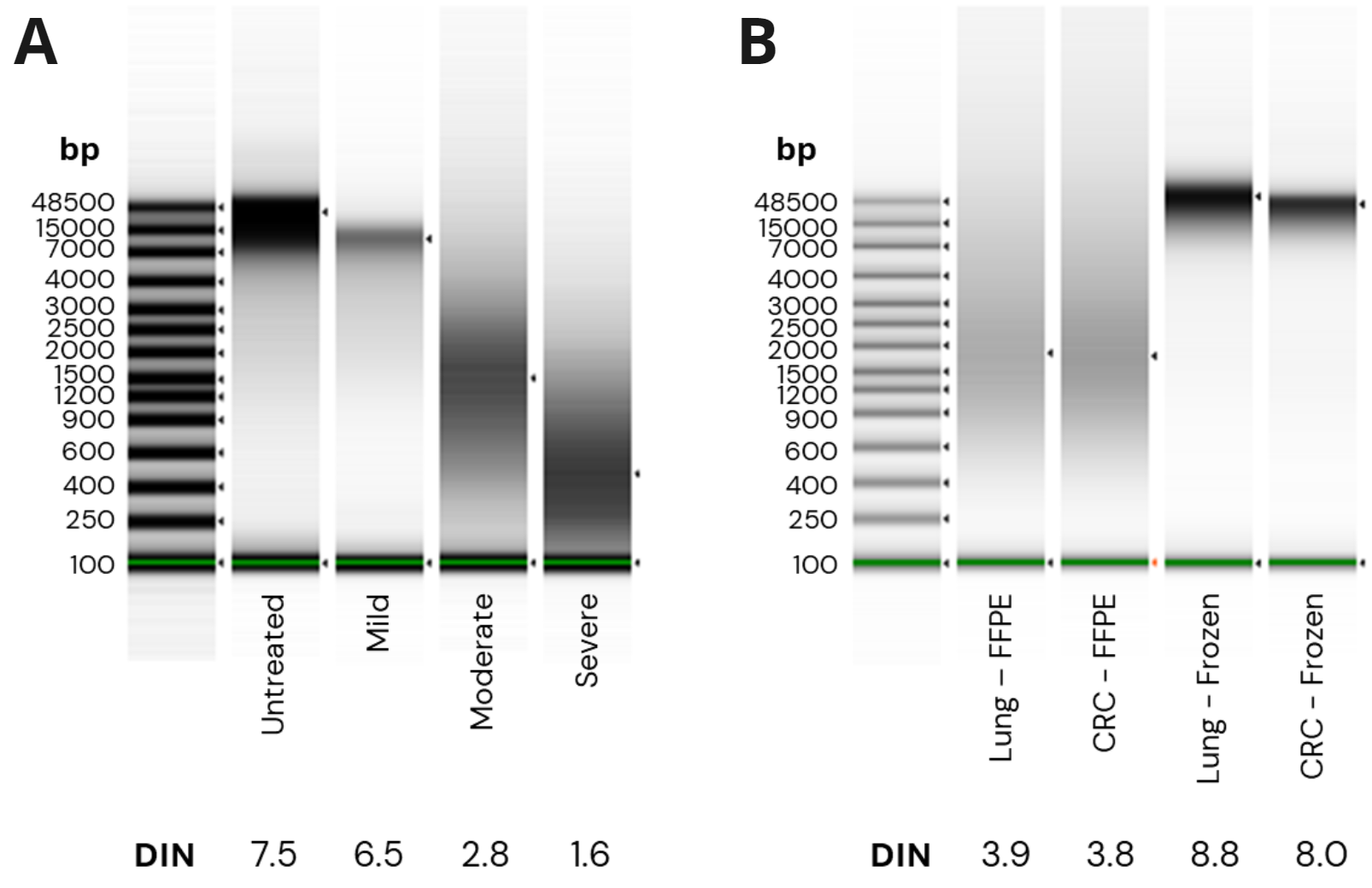
- fcDNA quality was assessed using Agilent Genomic DNA ScreenTape assay. Damage levels were mild, moderate and severe corresponding to DNA Integrity Number (DIN) ranges of ≥ 5.1, 2.5 – 5.0 and ≤ 2.0 respectively.
- TapeStation traces for FFPE cancer samples show moderate damage with DIN values of 3.9 and 3.8. Matched FF samples show DIN scores ≥ 8.
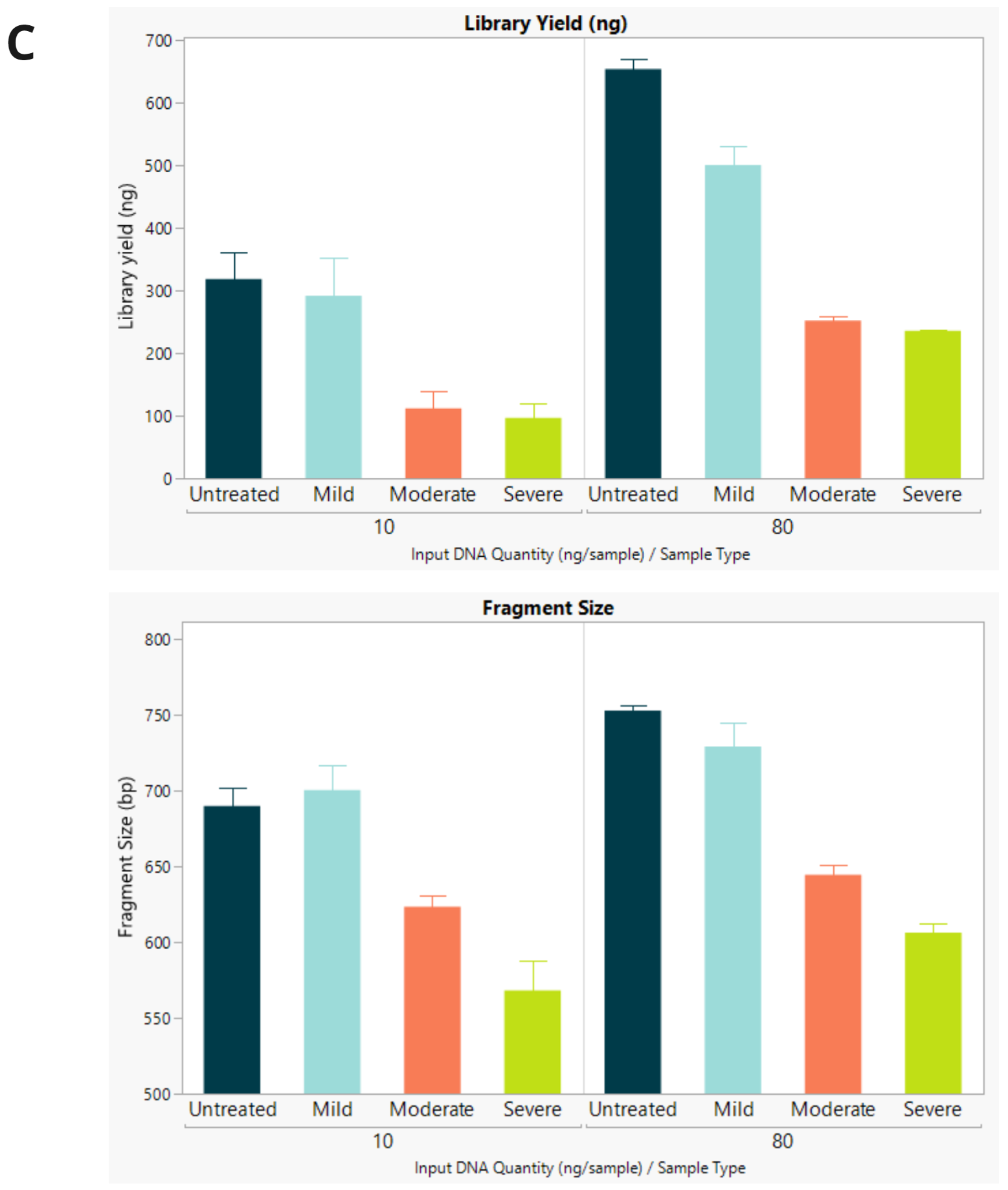
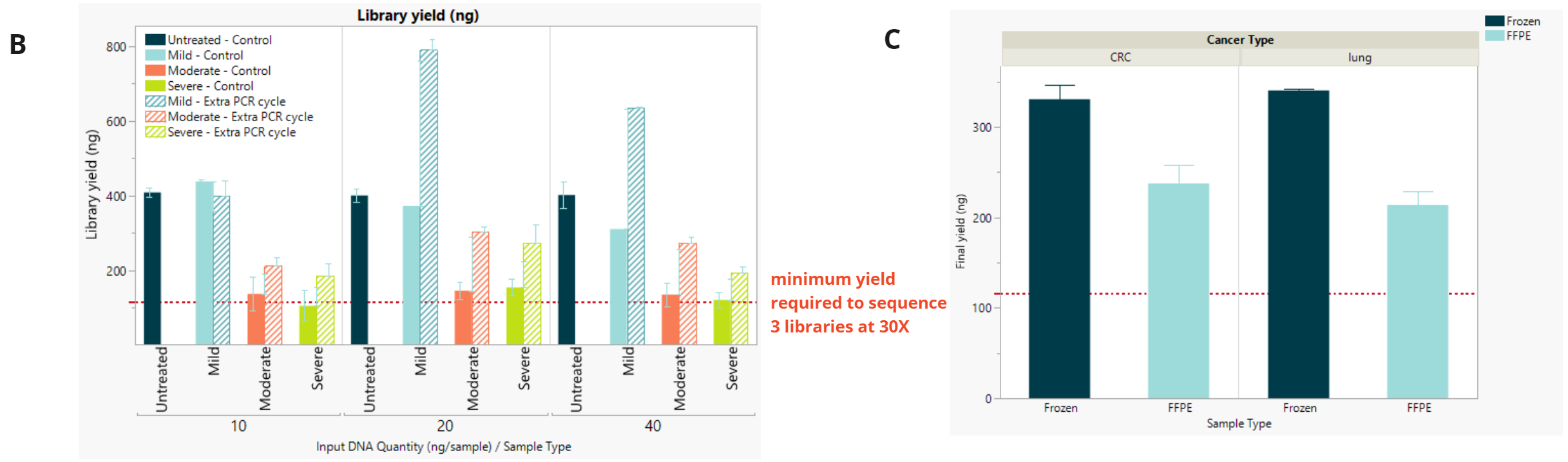
- Final library concentration and insert size for QMRS gDNA (untreated) and QMRS fcDNA (mild, moderate, severe). Increasing formalin damage reduced library yield and insert size.
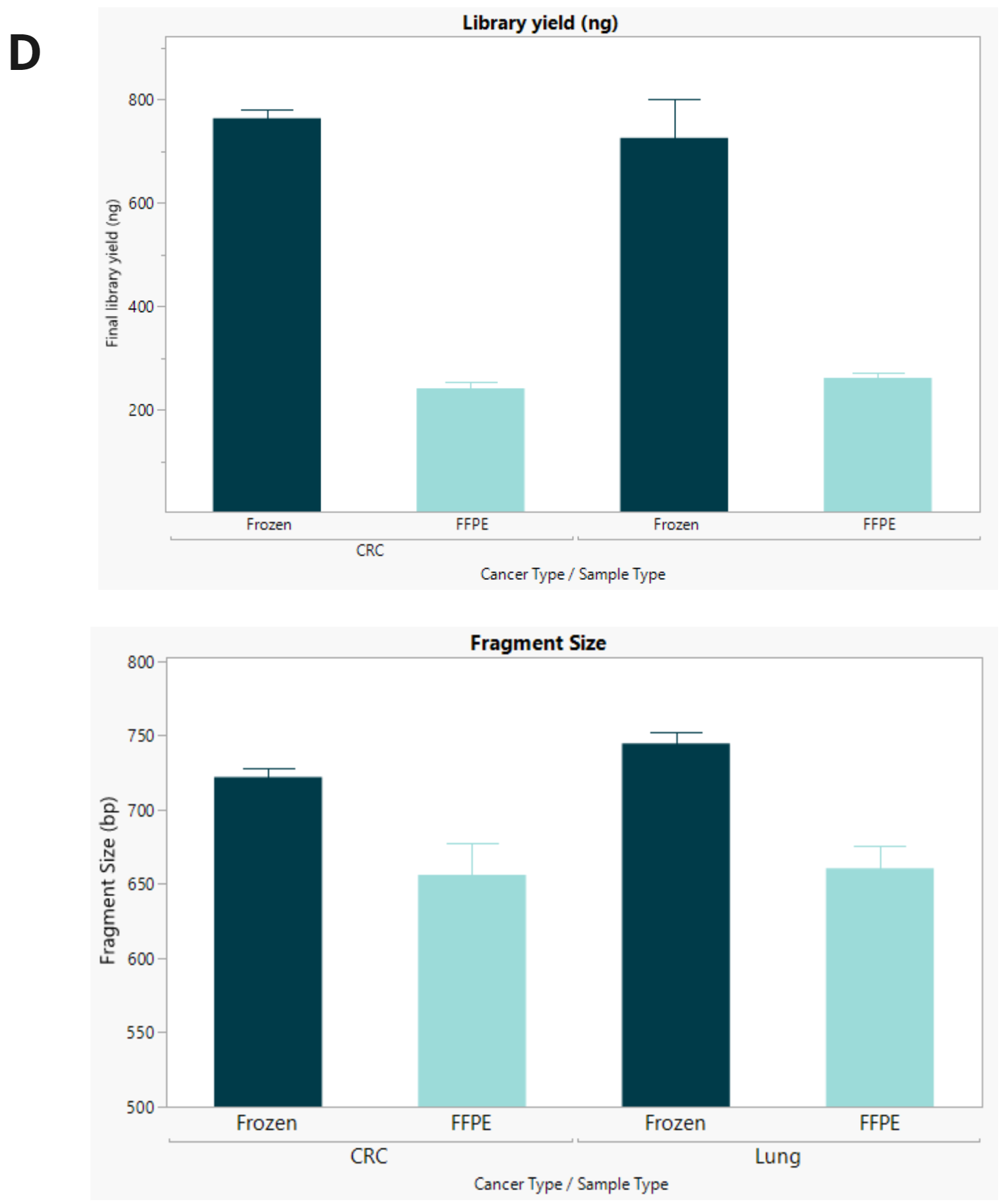
- Final library concentration and insert size for FFPE and FF samples. FFPE libraries have lower yield and insert size compared to matched FF samples.
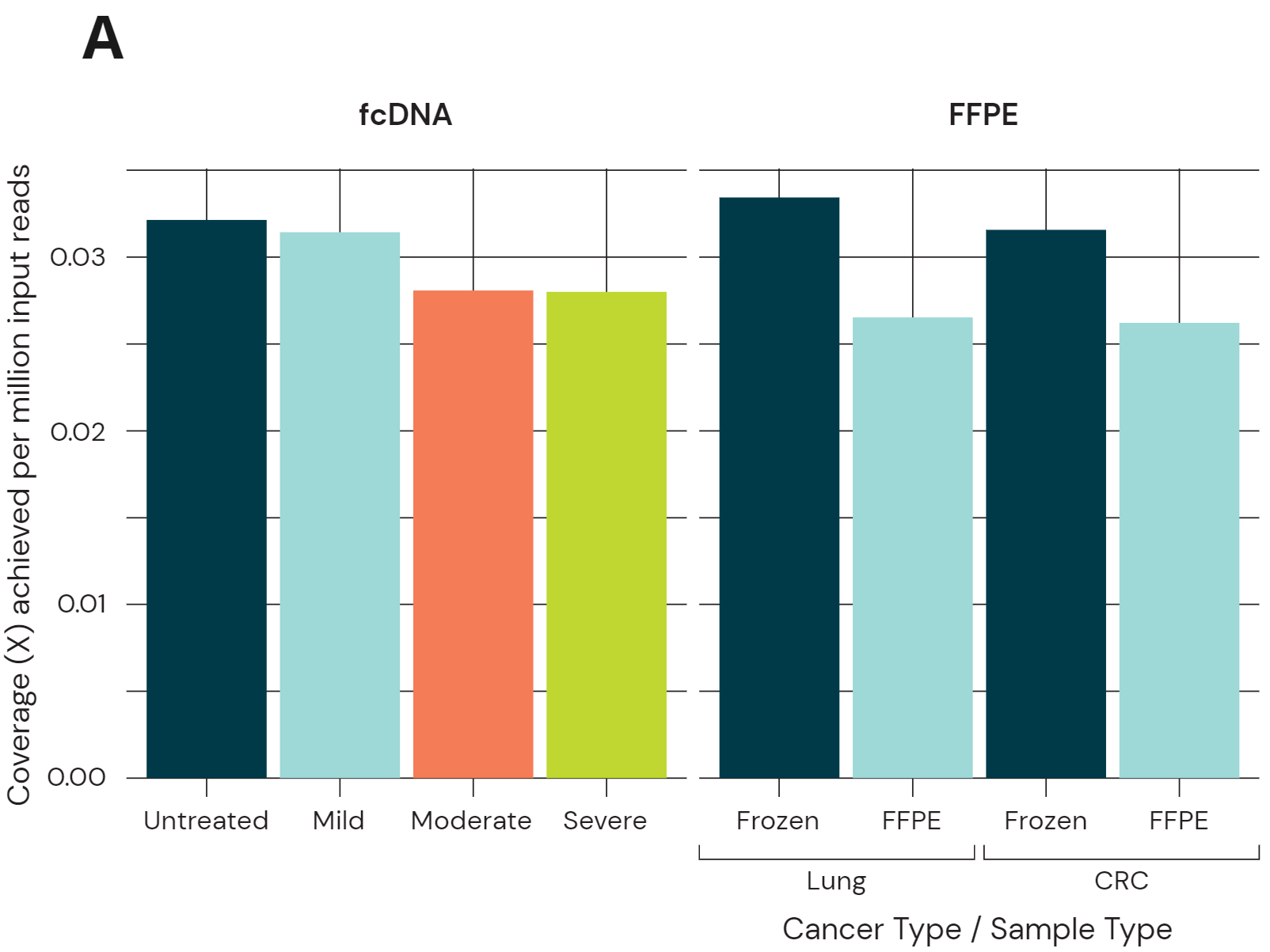
Sequencing performance using 80ng libraries of fcDNA and FFPE/Fresh Frozen. Libraries were sequenced on Illumina NovaSeq 6000 using a S4 Novaseq PE150 kit to ~30x coverage. Data was processed using biomodal’s duet analysis suite.
- Coverage yield normalised per million input reads. fcDNA resulted in a small drop in coverage which increases with more severe damage. FFPE cancer samples also show a small coverage drop, although still achieving acceptable coverage per million input reads for high depth sequencing.
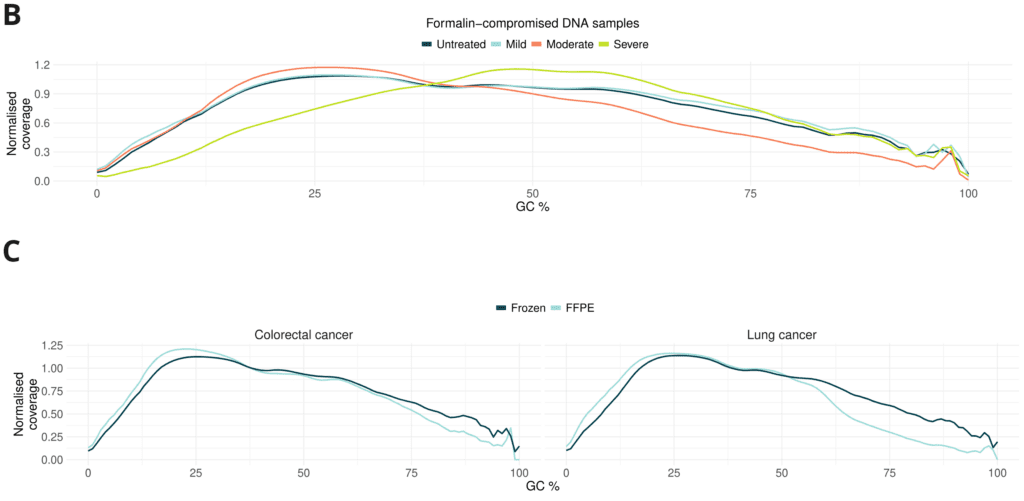
- Formalin damage only mildly alters GC coverage bias. GC coverage plots show similar distribution between untreated, mild and moderate. Severe damage resulted in reduced representation of low %GC content.
- CRC FFPE libraries show no significant difference compared to fresh frozen. Lung cancer FFPE libraries have slightly lower coverage at high GC content.


Epigenetic analysis of biomodal’s pipeline run on formalin-compromised DNA standards and FFPE samples.
- Correlation of 5mC levels at single CpG resolution (5mC) or averaging across 100kbp windows (5hmC) were measured between untreated control and fcDNA samples with different levels of formalin damage. All fcDNA samples show high mC correlation with Pearson’s r >0.9, including for the most severe formalin damage. Reduced correlation for 5hmC arises from the overall scarcity of this modification in these samples (<1%)
- Cancer samples also show high correlation between fresh frozen and FFPE-treated samples.
- Relative correlation levels for 5mC (single CpG) and 5hmC (100 kbp windows). 1 = average Pearson’s r for 4 NA12878 technical replicates.






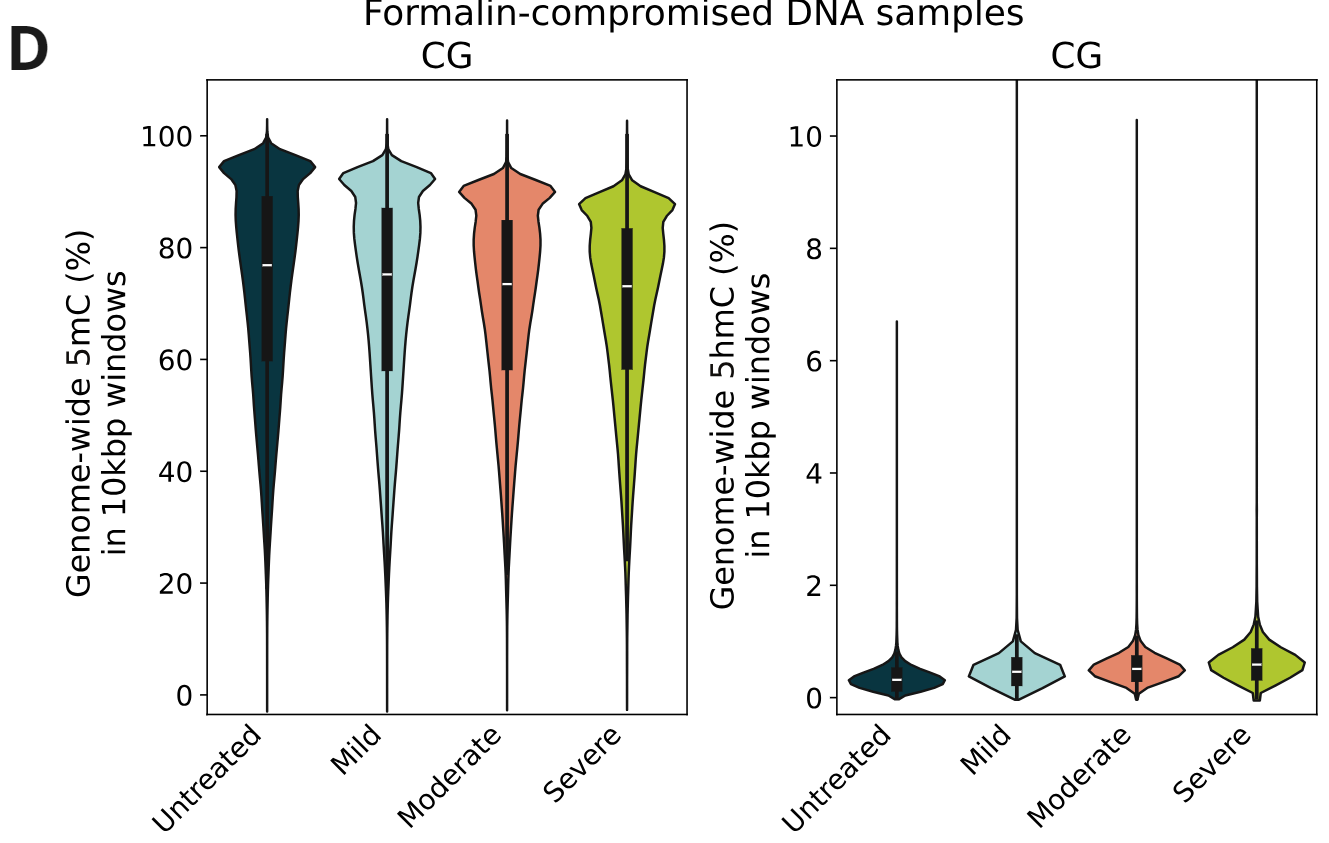
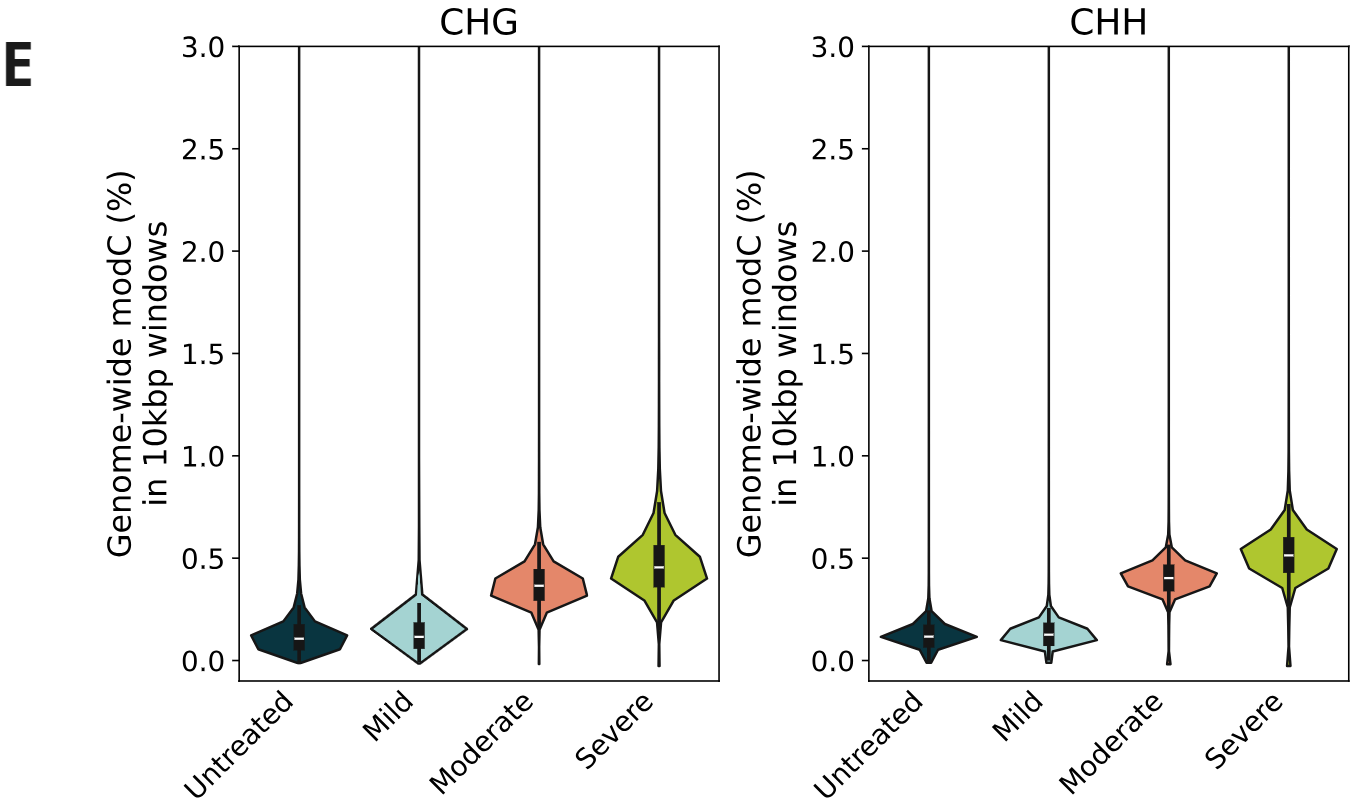
Genome-wide 5mC and 5hmC level analysis in CpG, CHG and CHH contexts.
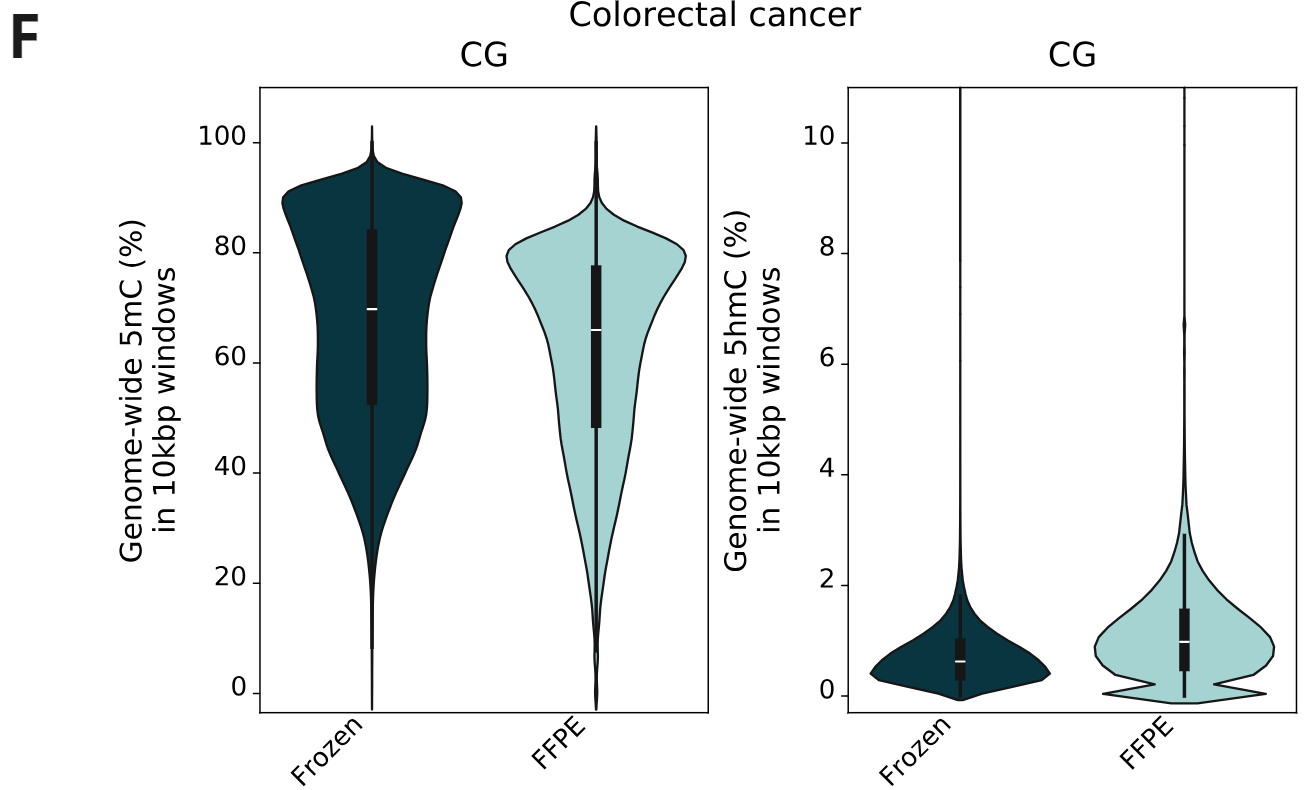
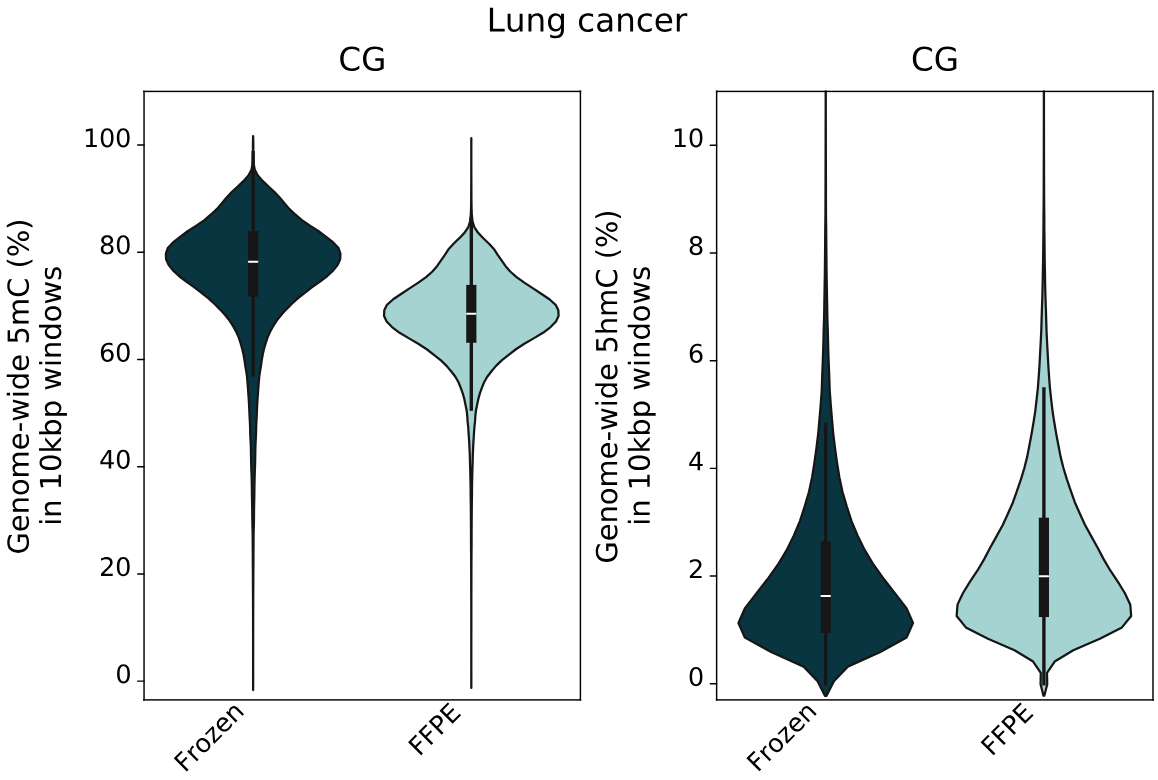
- Percentage of cytosines called as 5mC and 5hmC in CG contexts, calculated per 10kbp windows. 5mC levels decrease with increasing levels of formalin damage. Conversely, 5hmC levels show a small increase with formalin damage.
- Percentage of cytosines called as modC in CHG & CHH contexts, calculated per 10kbp windows. A small increase in modC levels associated with formalin damage is observed at non-CpG contexts.
- 5mC and 5hmC levels for fresh frozen and FFPE samples in CG contexts, calculated per 10kbp windows. The same trend as staged formalin damaged samples is observed, showing a genome-wide 5mC decrease and 5hmC increase.




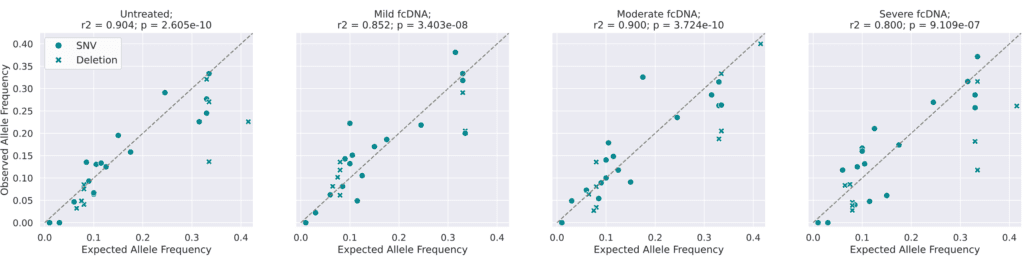
Analysis of variant allele frequencies (VAFs) on formalin-compromised DNA standards: Observed vs Expected Variant Allele Frequencies. Expected allele frequencies (as measured by droplet digital PCR) were obtained from Horizon(1) and observed allele frequencies were calculated by aligning reads to the reference genome and counting the number of reads supporting the variant as a fraction of the total coverage for that position.

| Standard PCR cycles | Recommended PCR cycles | Standard PCR cycles | Recommended PCR cycles | Standard PCR cycles | Recommended PCR cycles | |
|---|---|---|---|---|---|---|
| DNA input range | 10ng – <20ng | 20ng – <40ng | 40ng – 80ng | |||
| Untreated | 8 | N/A | 7 | N/A | 6 | N/A |
| fcDNA and FFPE | 8 | 9 | 7 | 8 | 6 | 7 |


Optimisation of library yield for damaged samples
Due to the decrease in final library yield observed with damaged fcDNA and FFPE samples, changes are recommended to the standard duet evoC workflow in order to generate enough library to use on an S4 flowcell at 30x coverage. For FFPE damaged libraries we propose to add one additional PCR cycle.
- duet evoC libraries were generated from fcDNA in duplicate from 10, 20 and 40ng input (minimum input for each range) using either recommended PCR cycles from the user guide or with 1 additional cycle.
- Library yield (ng) comparing standard number of PCR cycles (control) and additional cycle. For all inputs, libraries with an additional PCR cycle achieved enough yield to allow 3 sequencing runs at 30x coverage assuming a pooled library concentration of 2nM for a S4 NovaSeq PE150 kit (red dotted line).
- Library yield (ng) achieved for FFPE libraries at the minimum 10 ng input using 1 additional cycle to user guide (9 cycles for FFPE and 8 cycles for Fresh Frozen).
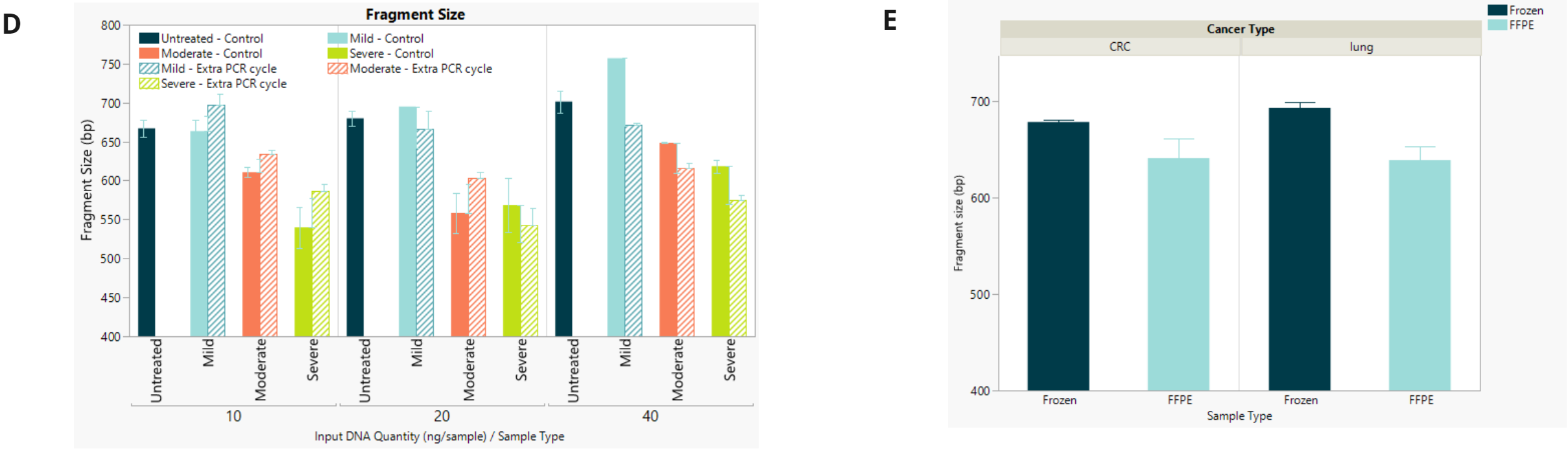
- Final library fragment size for fcDNA and FFPE shows expected drop regardless of the number of PCR cycles.
- Same as above.
In conclusion we demonstrate the compatibility of duet multiomics solution evoC with formalin-damaged DNA. While damage resulted in lower library yields and insert sizes relative to undamaged controls, high accuracy genetic and epigenetic base resolution data is produced even at severe levels of formalin-induced damage. We note small changes in modified cytosine calling dependent on sequence context: a reduction in 5mC and an increase in 5hmC for CpGs, and an increase of modC for CHH/CHG. We hypothesise this may be due to slightly different formalin-induced deamination pathways acting on unmodified cytosines (predominately found at CHH/CHG) and modified cytosines (much more prevalent in CpG contexts). Additionally, no effect on allele frequency detection was observed for the fcDNA QMRS standards.
Finally, for formalin-damaged DNA, we recommend increasing the number of PCR cycles (at least one additional cycle) to have sufficient library yield for deep sequencing.
- Simultaneous sequencing of genetic and epigenetic bases in DNA, Füllgrabe and Gosal et al., Nature Biotechnology (2023). (duet multiomics solution technology paper)
- Quantitative Multiplex Reference Standard (Horizon Discovery)
- Hedegaard, Jakob et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PloS one vol. 9,5 e98187. 30 May. 2014.

