duet multiomics solution evoC
Uncover a true 6-base genome for unprecedented insights into current and future biological states. Access all four canonical bases and distinguish 5‑methylcytosine (5mC) and 5‑hydroxymethylcytosine (5hmC) from a single 5ng DNA sample, in a single experiment, with high accuracy.
The information encoded within our DNA is not just held within the four bases – A, C, G and T – but also within the epigenetic modifications 5mC and 5hmC. Only with a 6-base genome will we achieve comprehensive genomic insight and truly understand the dynamic nature of the human genome.
duet multiomics solution evoC reveals a correlation in 5mC and 5hmC by distinguishing between the two within regions of open chromatin and gene expression. Researchers can use the data to build predictive models of gene expression, chromatin accessibility, and enhancer status, gaining unprecedented insights into current and future states of disease.

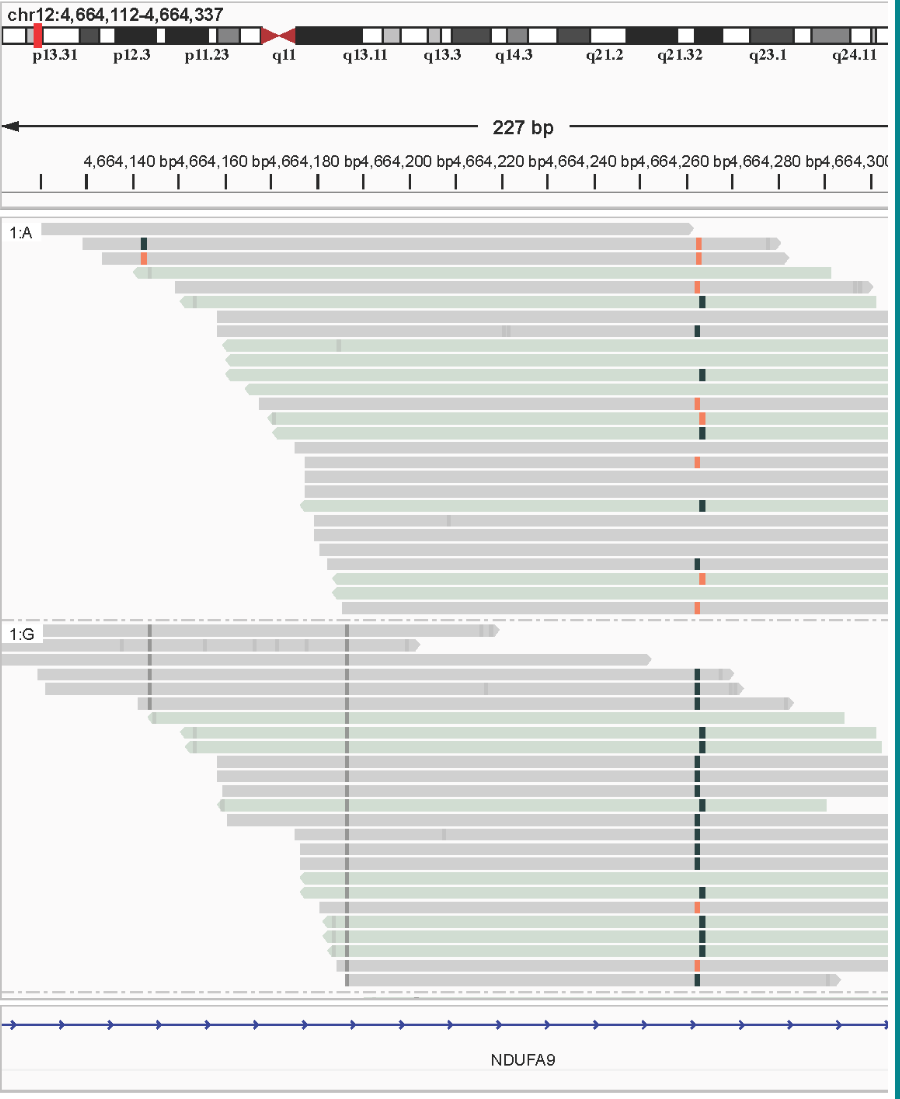
The unique ability to accurately determine genetic and epigenetic variation on the same read is a powerful feature of duet evoC data, and allows you to directly link genetic variants with methylation variation.
Here is an example of allelic variant-associated methylation and hydroxymethylation where a G→A germline variant favours either methylation or hydroxymethylation respectively. This is found in the NDUFS2 gene, which has demonstrated links to cancer and age-related diseases. Methylation is shown in black, and associated with the G allele. Hydroxymethylation is shown in red/orange, and associated with the A allele.
By distinguishing 5mC from 5hmC, duet evoC reveals a correlation in 5mC and 5hmC within regions of open chromatin and gene expression.
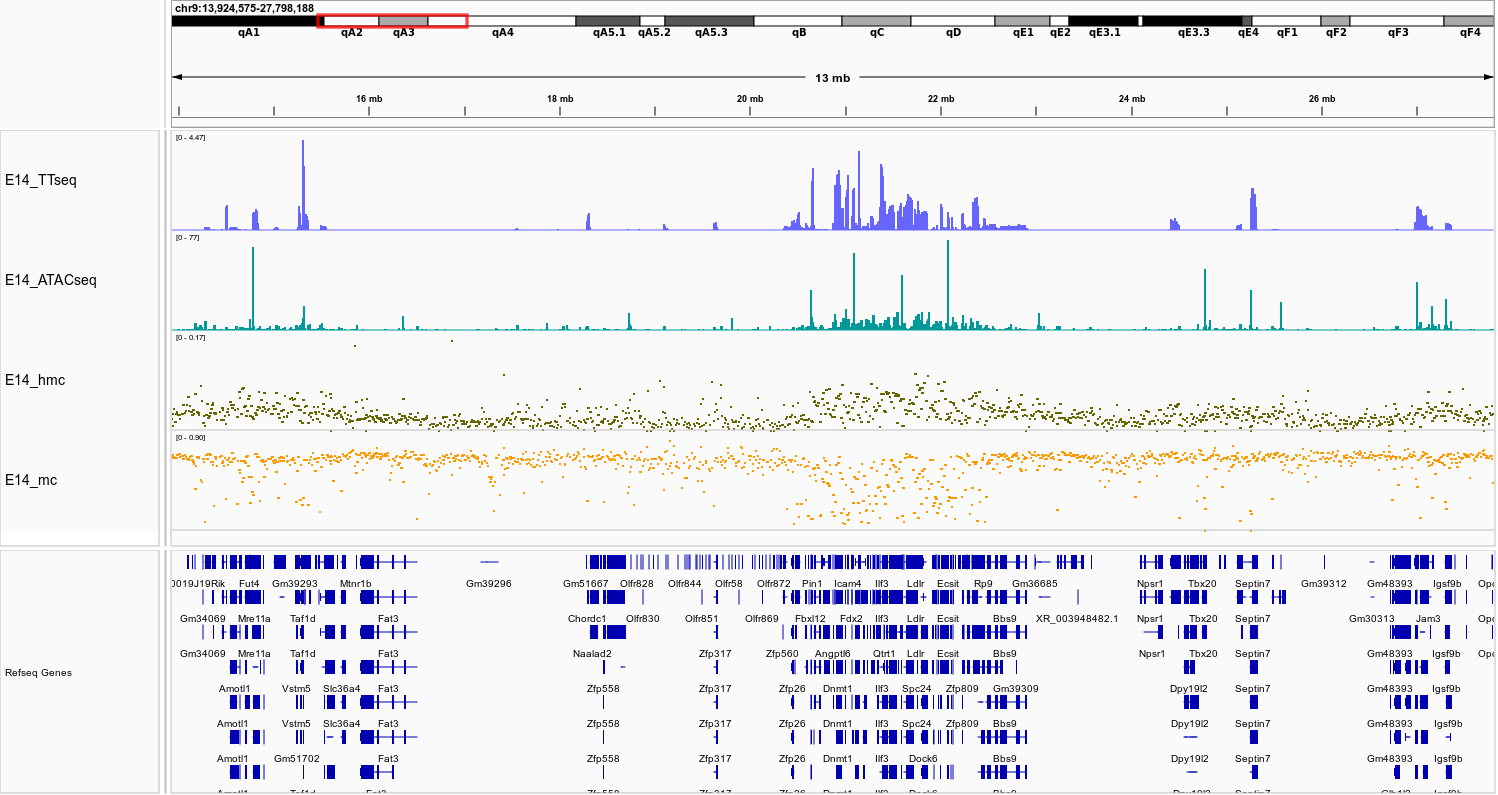
Changes in gene expression (blue) and chromatin accessibility (green) correlate with changes in 5hmC (brown) and 5mC (yellow) in E14 cells.
duet multiomics solution evoC combines the duet evoC library preparation assay and the duet software package to investigate genetic sequence and differentiated 5mC and 5hmC simultaneously.
The duet evoC library preparation kit takes a low input DNA sample (5ng) and copies each DNA strand across a hairpin structure. The original strand maintains both methylated and hydroxymethylated cytosine modifications, and is used for epigenetic analysis, and the copy strand is used for genetic sequence analysis.
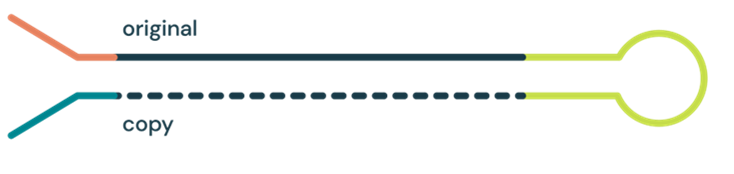
The duet software then takes the sequence information from pairs of original and copy strands and aligns them. For each position, it compares both original and copy strands to compute a resolved base that has complete genetic and epigenetic information.
From there, use our comprehensive data analysis package to explore methylation profiles, genetic variants, and variant-associated methylation in your samples, and compare epigenetic changes across genetic windows or contexts, in a single sample or within multi-sample cohorts.

| Unit | Product code |
|---|---|
duet multiomics solution evoC 8 reaction kitduet multiomics solution evoC contains:
|
6205 |
duet multiomics solution evoC 24 reaction kitduet multiomics solution evoC contains:
|
6206 |
duet UDIs: 1-8 reaction kit |
4101 |
duet UDIs: 1-24 reaction kit |
4102 |
duet UDIs: 1-96 reaction kit |
4104 |
Please contact us for orders or for more information
Find out how our solution can help you gain a deeper understanding of biology
The four traditional bases make up the foundational genetic code for all living systems. DNA methylation affects how and when genes are expressed, but the two methylation marks 5mC and 5hmC have opposing effects: 5mC reduces expression, or “turns off” genes, and in contrast, 5hmC increases gene expression, and “turns on” or “primes” genes. These two marks have unique functions, and capturing their distinct information, alongside traditional genetic information, is essential to understanding normal biology and disease states.
Our duet multiomics solution evoC protects cytosines that display epigenetic markers during library preparation without degrading your sample. 5mC markers are copied from the original DNA strand to a complimentary synthesized strand attached with a hairpin sequence, whilst 5hmC markers remain only on the original strand. Unmodified cytosines on both strands are then enzymatically deaminated. The duet bioinformatics pipeline uses base information from both strands to decode the original base at each position along with epigenetic marks highlighted as the fifth and sixth genetic letter, resulting in a 6-base genome. For more information on how this occurs, read our paper here.
No, the duet multiomics solution evoC integrates seamlessly with your current Illumina sequencer and bioinformatics pipeline.
Our workflow is similar to other standard library preparation and sequencing workflows, with an assay turnaround time of less than 18 hours and an assay hands-on time of less than 10 hours.