We present a computational toolkit to analyse 5mC and 5hmC modifications at scale and describe its performance on a novel liquid biopsy dataset.
Methylation data has diverse applications in cancer, including early-stage diagnosis through liquid biopsy, classification to guide treatment pathways, and prognosis. However, analyzing methylation data poses significant challenges, as it is constrained by scalability and usability issues.
Our recently introduced technology, duet multiomics solution evoC, enables the reading of 6-base information (A, T, G, C, 5mC and 5hmC) from DNA, further amplifying the complexity and scale of datasets generated in a single sequencing experiment. To address this, we present fast and scalable array-based analysis software (called modality) for the analysis of 6-base genomes, using multi-core out-of-memory processing to enable extremely efficient computation, even for datasets that are too large to fit into memory.
Unlike existing tools that exceed typical laptop memory with ~10 samples, the modality analysis software can efficiently analyse (e.g. DMR calling) a colorectal cancer liquid biopsy dataset of over 100 samples in minutes on a standard laptop.
The software combines efficient computation with tools for exploratory (e.g., plotting, methylation summaries) and downstream analyses (e.g., DMR identification, PCA). Designed for efficiency and ease of use, it enables users to rapidly transition from raw data to actionable insights and publication-ready visualisations. As multiomic data become the standard in cancer research, our data structure supports the future integration of additional data types, with the promise of enabling combined genomic and epigenomic data from solutions like duet evoC. This will enable streamlined and efficient multiomic analysis to uncover deeper biological insights.
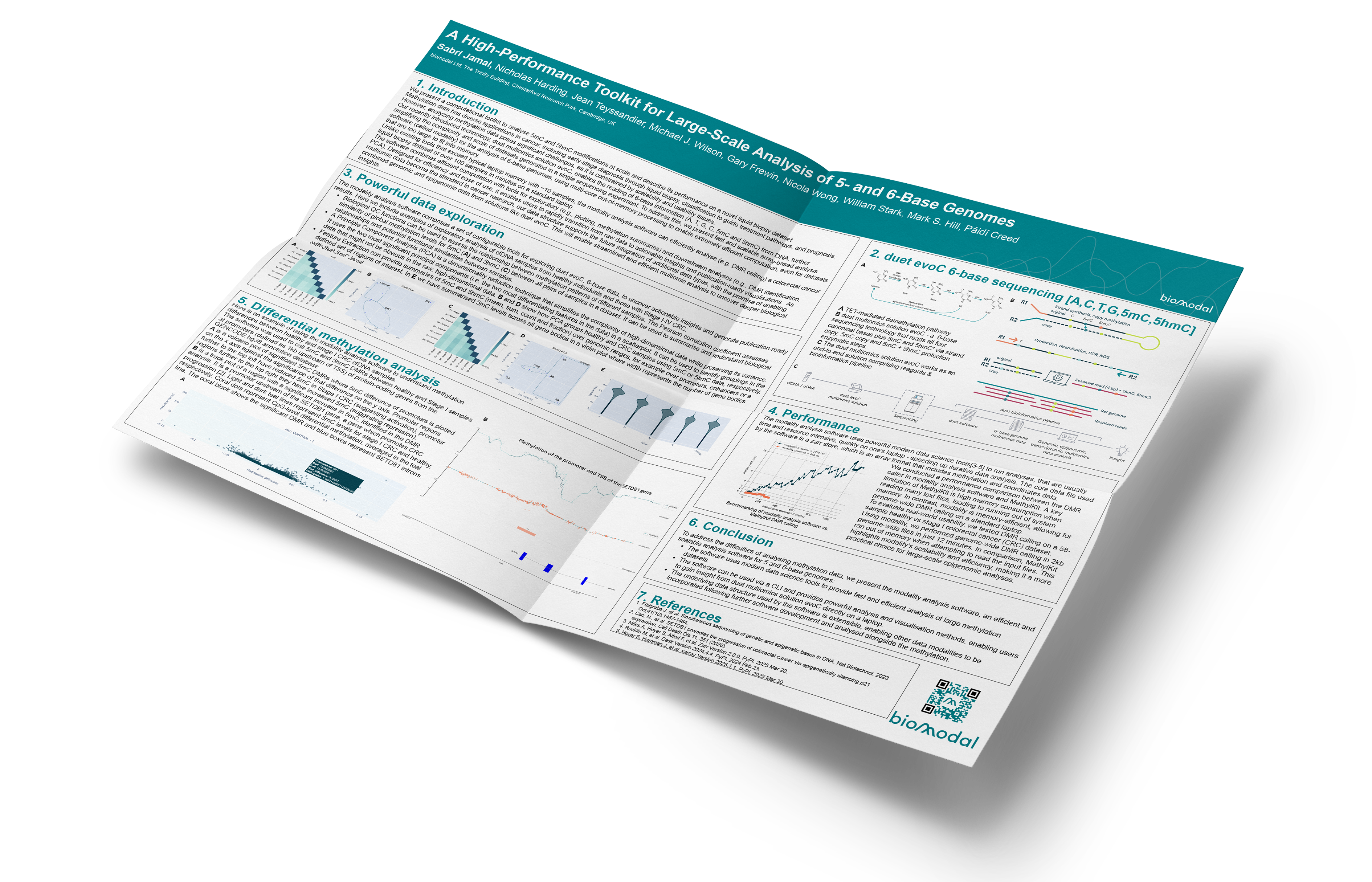
A TET-mediated demethylation pathway
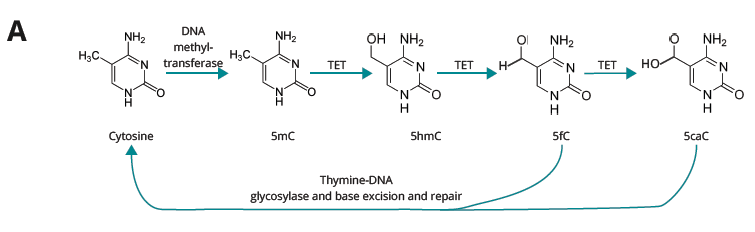
B duet multiomics solution evoC – a 6-base sequencing technology that reads all four canonical bases plus 5mC and 5hmC¹ via strand copy, 5mC copy and 5mC + 5hmC protection enzymatic steps.
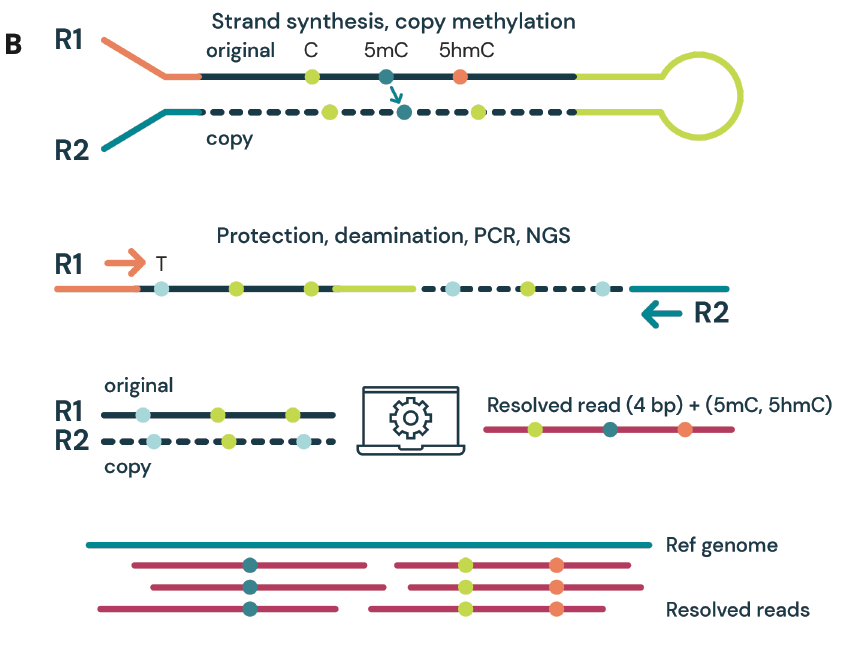
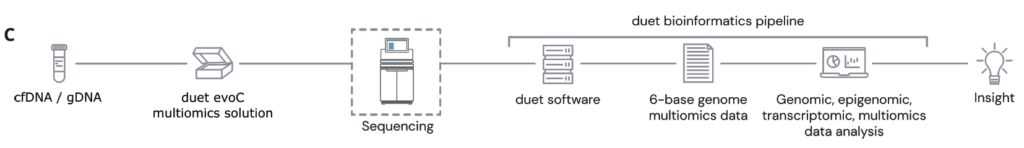
C The duet multiomics solution evoC works as an end-to-end solution comprising reagents & bioinformatics pipeline
The modality analysis software comprises a set of configurable tools for exploring duet evoC, 6-base data, to uncover actionable insights and generate publication-ready results. Here we include examples of exploratory analysis of cfDNA samples from healthy individuals and those with Stage I-IV CRC:
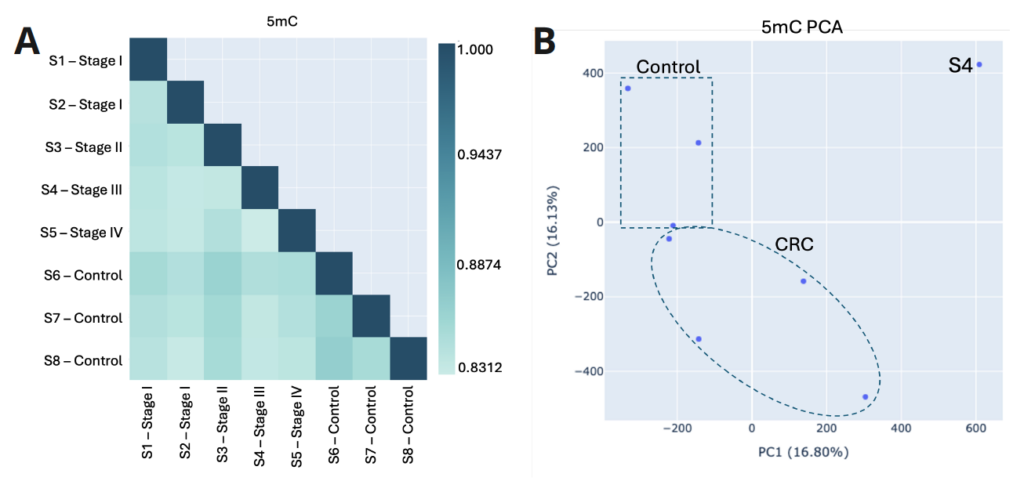
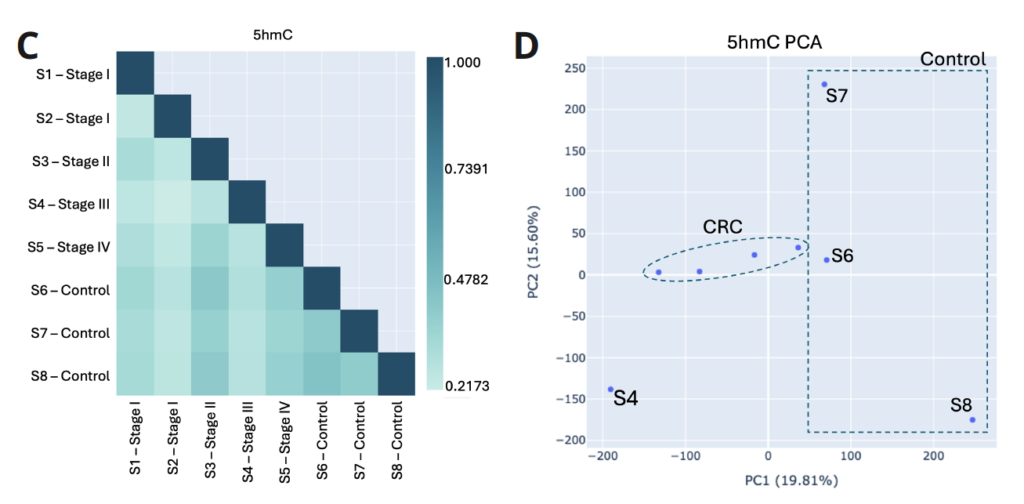
- Biological QC functions can be used to assess the relationship between methylation patterns of different samples. The Pearson correlation coefficient assesses similarity of global methylation levels for 5mC (A) and 5hmC (C) between all pairs of samples in a dataset. It can be used to summarise and understand biological relationships and potential functional similarities between samples.
- A Principle Component Analysis (PCA) is a dimensionality reduction technique that simplifies the complexity of high-dimensional data while preserving its variance. It uses the two most significant principal components (i.e. the two most differentiating features in the data) in a scatterplot. It can be used to identify groupings in the data that might not be obvious in the raw, high-dimensional data. B and D show how PCA groups healthy and CRC samples using 5mC or 5hmC data, respectively.
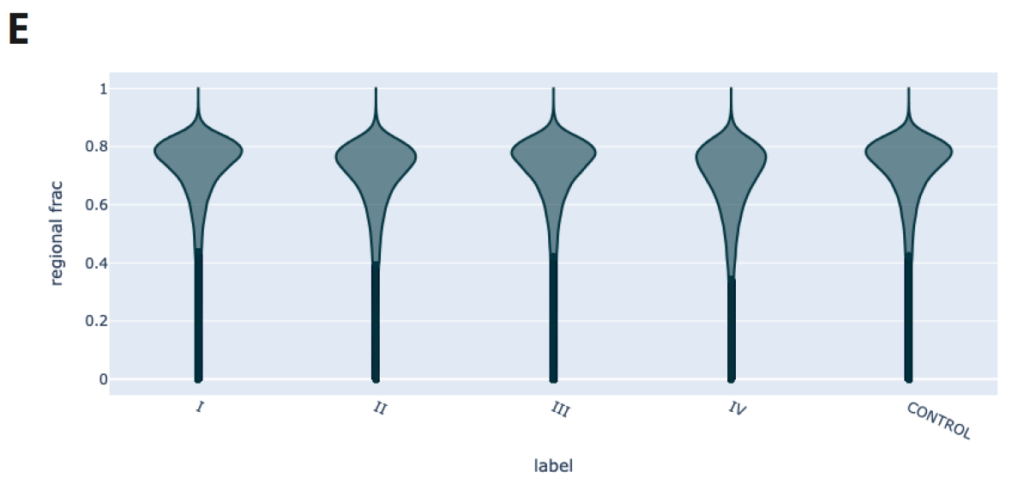
- Feature Extraction can provide summaries of 5mC and 5hmC (mean, sum, count and fraction) over genomic ranges, for example over promoters, enhancers or a defined set of regions of interest. In E we have summarised 5mC levels across all gene bodies in a violin plot where width represents the number of gene bodies with that 5mC level.
The modality analysis software uses powerful modern data science tools[3-5] to run analyses, that are usually time and resource intensive, quickly on one’s laptop – speeding up iterative data analysis. The core data file used by the software is a zarr store, which is an array format that includes methylation and coordinates data.
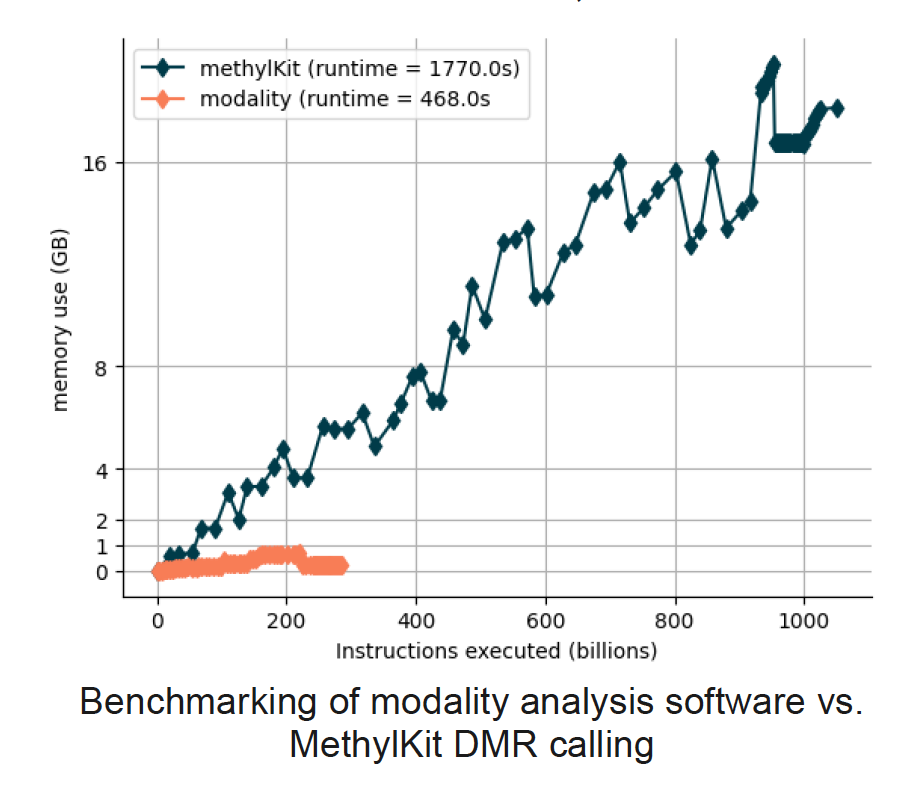
We conducted a performance comparison between the DMR caller in modality analysis software and MethylKit. A key limitation of MethylKit is high memory consumption when reading many text files, leading to running out of system memory. In contrast, modality is memory-efficient, allowing for genome-wide DMR calling on a standard laptop.
To evaluate real-world usability, we tested DMR calling on a 58-sample healthy vs stage I colorectal cancer (CRC) dataset. Using modality, we performed genome-wide DMR calling in 2kb genome-wide tiles in just 12 minutes. In comparison, MethylKit ran out of memory when attempting to read the input files. This highlights modality’s scalability and efficiency, making it a more practical choice for large-scale epigenomic analyses.
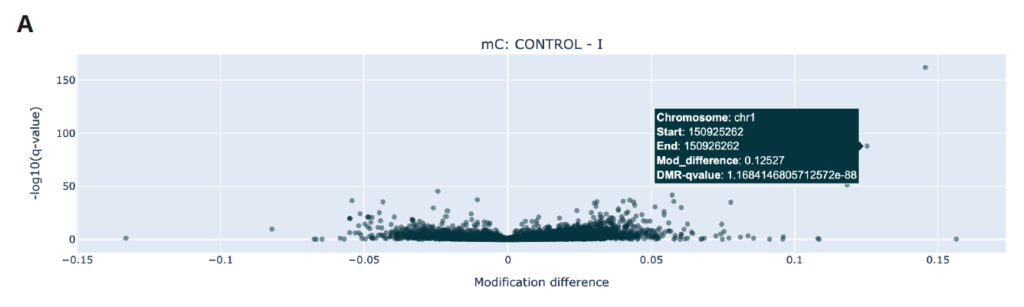
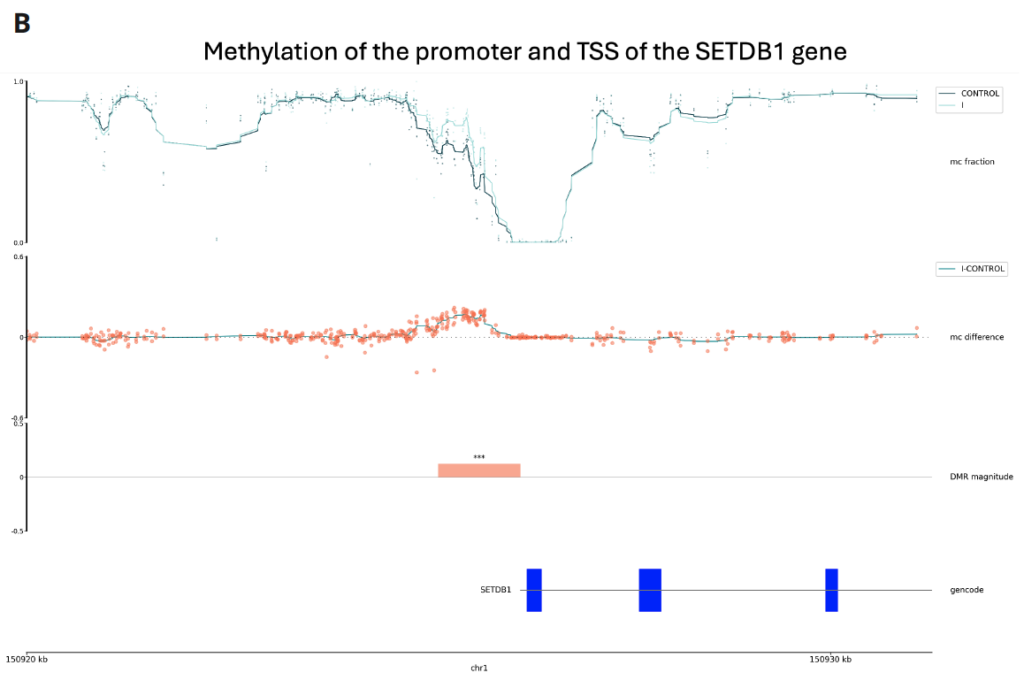
Here is an example of using the modality analysis software to understand methylation differences between healthy and stage I CRC cfDNA samples.
The software was used to call 5mC and 5hmC DMRs between healthy and Stage I samples at promoters (defined as 1kb upstream of TSS) of protein-coding genes from the GENCODE hg38 annotation database.
A is a volcano plot of significant 5mC DMRs where 5mC difference of promoters is plotted on the x axis against the significance of that difference on the y axis. Promoter regions further to the top left have reduced 5mC in Stage I CRC (suggesting activation), promoter regions further to the top right they have increased 5mC (suggesting repression).
B is a track plot of a region with a significant increase in 5mC, identified in the DMR analysis. It is a promoter upstream of the SETDB1 gene, a gene which promotes CRC progression [2]. Light and dark teal lines represent 5mC levels for stage I CRC and healthy, respectively. Coral dots represent CpG-level differential methylation, averaged in the teal line. The coral block shows the significant DMR and blue boxes represent SETD81 introns.
To address the difficulties of analysing methylation data, we present the modality analysis software, an efficient and scalable analysis software for 5 and 6-base genomes:
- The software uses modern data science tools to provide fast and efficient analysis of large methylation datasets.
- The software can be used via a CLI and provides powerful analysis and visualisation methods, enabling users to gain insight from duet multiomics solution evoC directly on a laptop.
- The underlying data structure used by the software is extensible, enabling other data modalities to be incorporated following further software development and analysed alongside the methylation.
- Füllgrabe J. et al. Simultaneous sequencing of genetic and epigenetic bases in DNA. Nat Biotechnol. 2023 Oct;41(10):1457-1464.
- Cao, N., et al. SETDB1 promotes the progression of colorectal cancer via epigenetically silencing p21 expression. Cell Death Dis 11, 351 (2020).
- Miles A, Hoyer S, Alted F, et al. Zarr Version 2.0.0. PyPI. 2025 Mar 20.
- Rocklin M, et al. Dask Version 2024.4.4. PyPI. 2024 Feb 23.
- Hoyer S, Hamman J, et al. xarray Version 2025.1.1. PyPI. 2025 Mar 30.