Discover the combinatorial power of genetics and epigenetics at single base resolution. We are delighted to present BaseSpace Sequence Hub data for biomodal’s duet multiomics solution +modC.
Genetics and epigenetics both offer important layers of information, but current technological methods make the integration of the two difficult and time-consuming with significant information loss. The duet multiomics solution +modC from biomodal achieves simultaneous genetic and DNA methylation sequencing data at single base resolution from the same DNA fragment, all in a single integrated workflow. This is achieved with minimal input DNA requirements and avoids the use of harsh chemicals that damage precious samples. The duet bioinformatics workflow performs comprehensive multiomic data analysis to report on all 4 canonical bases as well as methylated cytosine modifications, then integrates these two informational layers to give variant-associated methylation (VAM).
The duet multiomics workflow
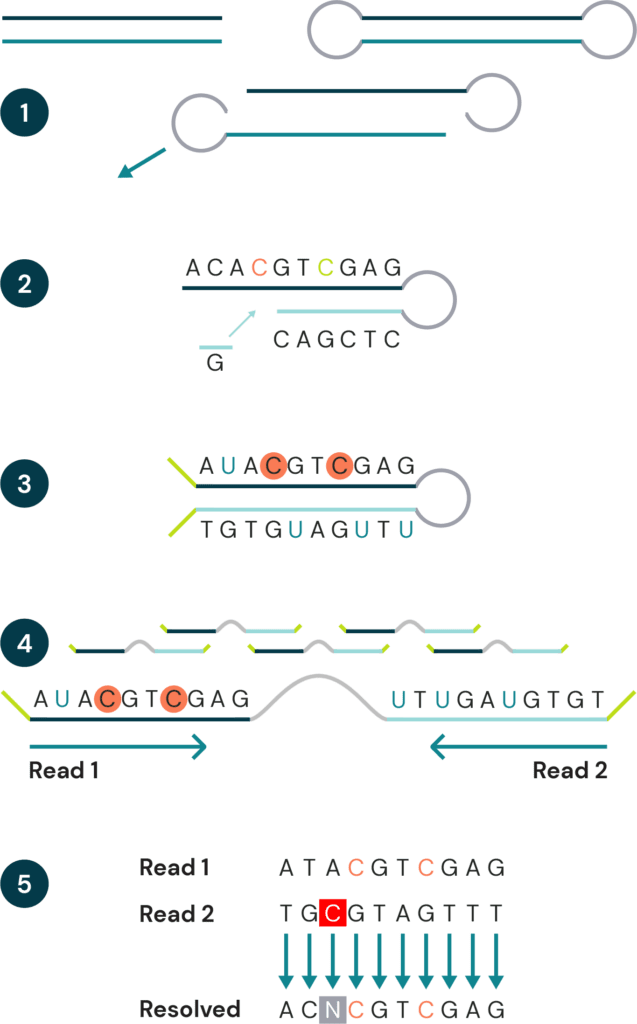
- Fragmented genomic DNA is ligated to a hairpin adapter at both ends. The hairpin complex is then split into two strands. Both strands move ahead to step 2 (one strand has been omitted for clarity)
- A complementary copy strand is synthesized, accurately capturing the genetic sequence prior to conversion.
- Modified cytosines are protected, and unmodified cytosines are deaminated to uracil. Sequencing adapters are ligated onto strand ends.
- The hairpin complex is linearised and PCR amplified, then sequenced. Read 1 sequences the original strand, read 2 sequences the copy strand. Uracil will be read as thymine.
- Post-sequencing, complementary reads are pairwise aligned. Bases from both reads are resolved using bespoke biomodal pipeline resolution rules to give the original base, plus cytosine modifications, for each position. Sequencing or PCR errors will result in impossible base pairings. These are identified, labelled as N, and filtered out.
To generate this sample data set, seven “Genome In A Bottle” samples were prepared using the biomodal duet multiomics solution +modC kit using 80ng DNA input each, and then sequenced in duplicate at 2×151 on NovaSeq 6000 S4. Sequencing data was processed using the biomodal pipeline v1.1.1 and downsampled to ~30X coverage.
The biomodal duet software pipeline takes fastQ files directly generated by the sequencer and performs a set of bioinformatic computational steps to produce analysis-ready data in industry-standard file formats. Genetic sequence alignment, methylation quantification, and variant calling are all reported as standard. Additionally, and unique to the duet multiomics solution, Variant-Associated Methylation information is accessible including an Allele-Specific Methylation file.
The following files and reports have been included for download:
- Summary_report: an Excel sheet containing summary metrics for all samples analysed. Metrics are associated with reads aligning to the reference genome, and with reads aligning to the spike-in controls. The sheet contains common NGS workflow metrics and duet multiomics solution +modC specific metrics.
- MultiQC: an HTML report including plots of the most important metrics for all samples including coverage, GC content and methylation bias.
- Allele_specific_methylation: Methylation on reads associated with each allele is quantified and a call of ASM is provided where asymmetry is significant.
- Genome_modC_quantification: files reporting methylated cytosine content for each sample across multiple genomic contexts (CpG, CHG, CHH)
- Variant_call_files: VCF file containing information about variants detected at specific positions in the reference genome.
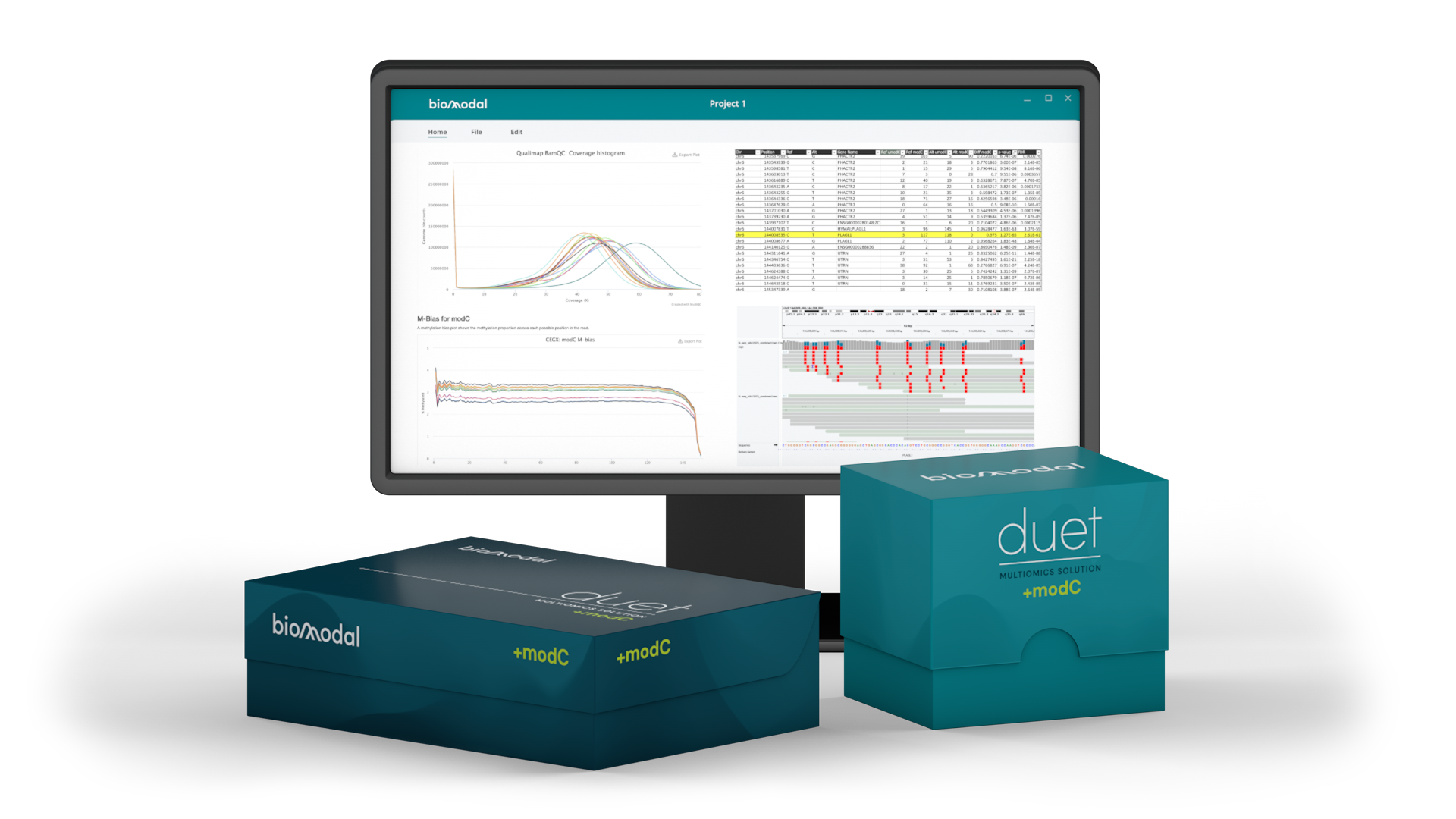
Example report
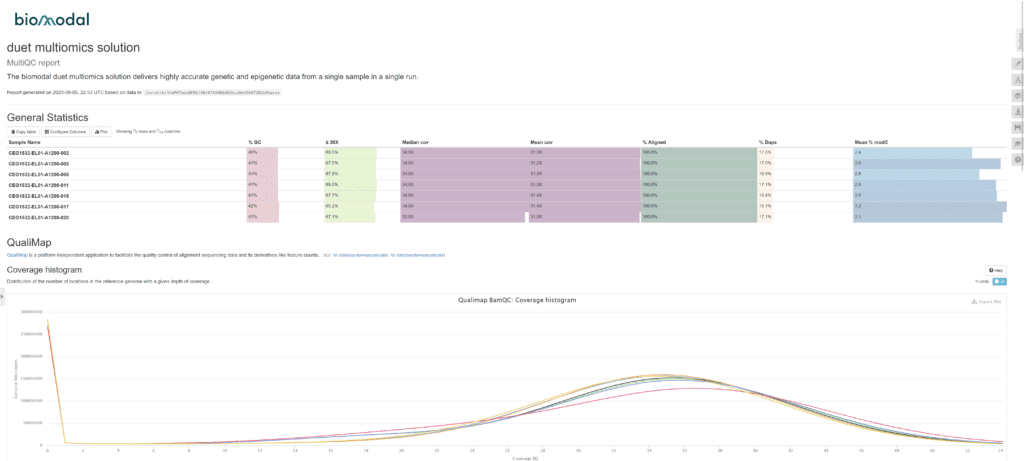
This example screenshot shows some of the information that the QC report contains, including coverage, %GC, and methylation fraction. The interactive HTML report offers these and a range of other important metrics in simple graphical formats, customisable for your research requirements.
Accessing Basespace
To view the data yourself, you can now import the project directly into your Basespace environment.