- Jean Teyssandier
- Walraj S. Gosal
- Jens Fullgrabe
- Nick Harding
- Riccha Sethi
- Michael Wilson
- Robert Crawford
- Ermira Lleshi
- Paula Golder
- Lisabet Andreasen
- Michael Hodgson
- Aurel Negrea
- Helen Sansom
- Ankita Singhal
- Minna Taipale
- Mengjie Li
- Yang Liu
- Alexandra Palmer
- Nikolay Pchelintsev
- Lidia Prieto-Lafuente
- Audrey Vandomme
- Gary Yalloway
- Páidí Creed
The predominant cytosine modification in DNA is methylcytosine (5mC). This base appears to exert a profound impact on gene expression patterns and is commonly linked to transcriptional repression. The introduction of hydroxymethylation (5hmC) adds a new layer of complexity to the conventional comprehension of DNA methylation dynamics, which is often thought to correlate with gene expression. Constrained to measuring four states of information, existing NGS-based technologies sacrifice genetic information for modification calling without distinguishing between these two important modification states. Genetic and methylation data combined together presents a unique opportunity to measure these cytosine modifications against gene expression.
duet multiomics solution evoC is a new sequencing technology that resolves all four genetic bases alongside the ability to distinguish the modification status of cytosines, discriminating 5mC from 5hmC (6 base calling). The technology consists of pre-sequencing library prep with enzymatic conversion of DNA together with an analysis pipeline, achieving base resolution of genetics and epigenetics at high accuracy. Here we present the method, alongside a high depth sequencing analysis of mouse embryonic stem cells. We demonstrate the potential of epigenetic modifications encoded in DNA to predict other important potential modulators of gene expression such as open chromatin and enhancer states as well as RNA expression itself.

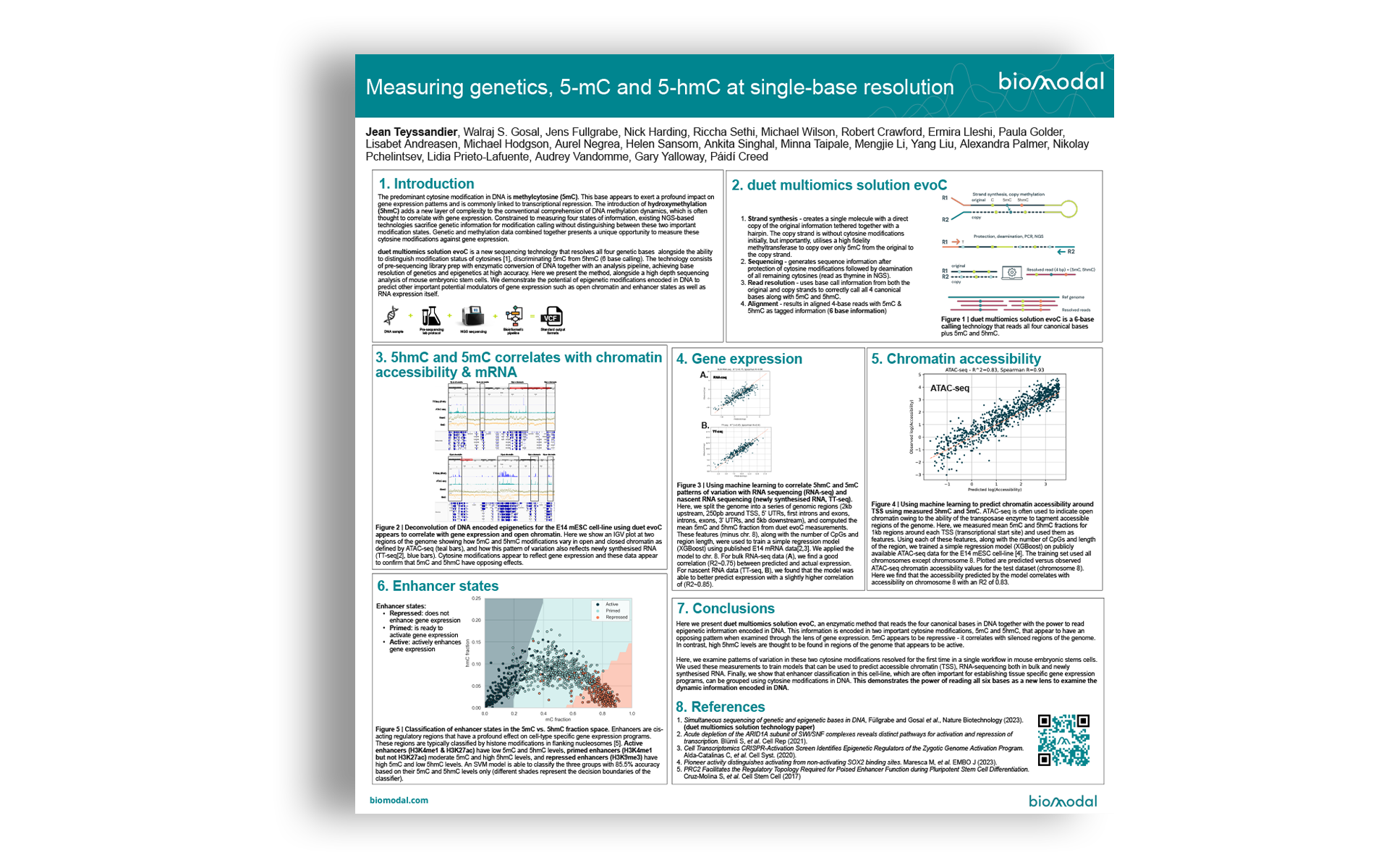
- Strand synthesis: creates a single molecule with a direct copy of the original information tethered together with a hairpin. The copy strand is without cytosine modifications initially, but importantly, utilises a high fidelity methyltransferase to copy over only 5mC from the original to the copy strand.
- Sequencing – generates sequence information after protection of cytosine modifications followed by deamination of all remaining cytosines (read as thymine in NGS).
- Read resolution – uses base call information from both the original and copy strands to correctly call all 4 canonical bases along with 5mC and 5hmC.
- Alignment – results in aligned 4-base reads with 5mC & 5hmC as tagged information (6 base information)
duet multiomics solution evoC is a 6-base calling technology that reads all four canonical bases plus 5mC and 5hmC.
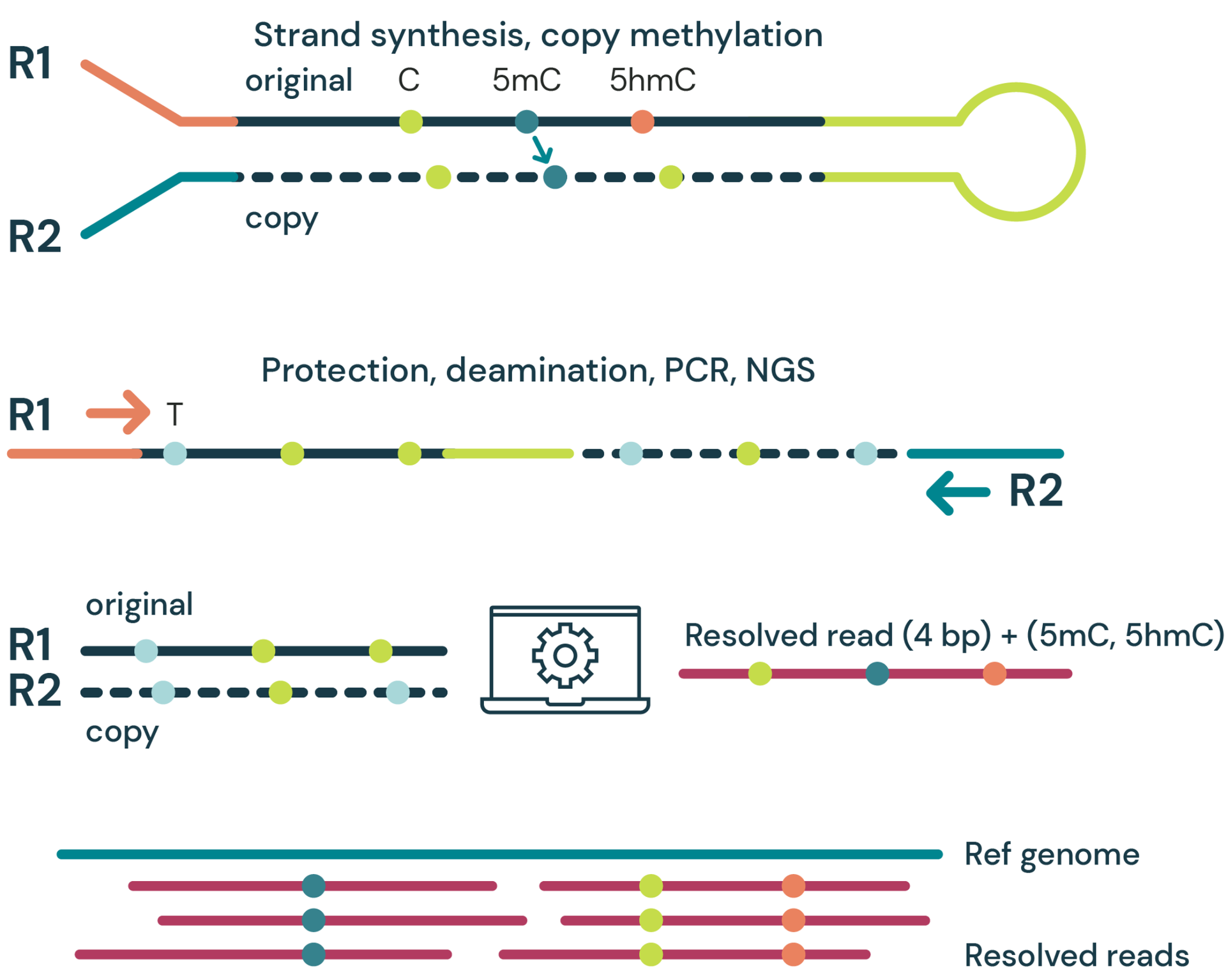
Deconvolution of DNA encoded epigenetics for the E14 mESC cell-line using duet evoC appears to correlate with gene expression and open chromatin.
Here we show an IGV plot at two regions of the genome showing how 5mC and 5hmC modifications vary in open and closed chromatin as defined by ATAC-seq (teal bars), and how this pattern of variation also reflects newly synthesised RNA (TT-seq, blue bars). Cytosine modifications appear to reflect gene expression and these data appear to confirm that 5mC and 5hmC have opposing effects.
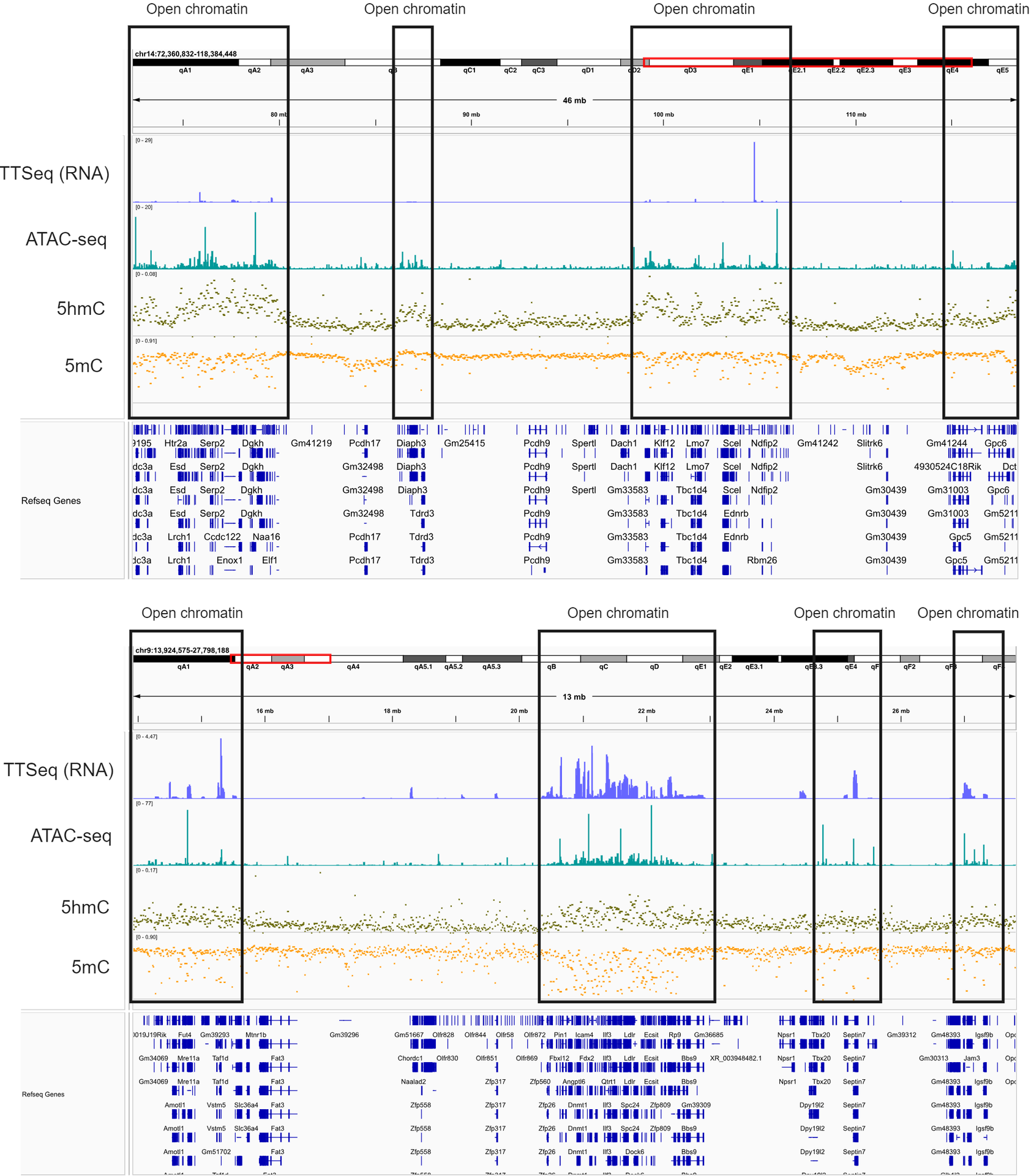
Using machine learning to correlate 5hmC and 5mC patterns of variation with RNA sequencing (RNA-seq) and nascent RNA sequencing (newly synthesised RNA, TT-seq).
Here, we split the genome into a series of genomic regions (2kb upstream, 250pb around TSS, 5′ UTRs, first introns and exons, introns, exons, 3′ UTRs, and 5kb downstream), and computed the mean 5mC and 5hmC fraction from duet evoC measurements. These features (minus chr. 8), along with the number of CpGs and region length, were used to train a simple regression model (XGBoost) using published E14 mRNA data.
We applied the model to chr. 8. For bulk RNA-seq data (A), we find a good correlation (R2~0.75) between predicted and actual expression.
For nascent RNA data (TT-seq, B), we found that the model was able to better predict expression with a slightly higher correlation of (R2~0.85).
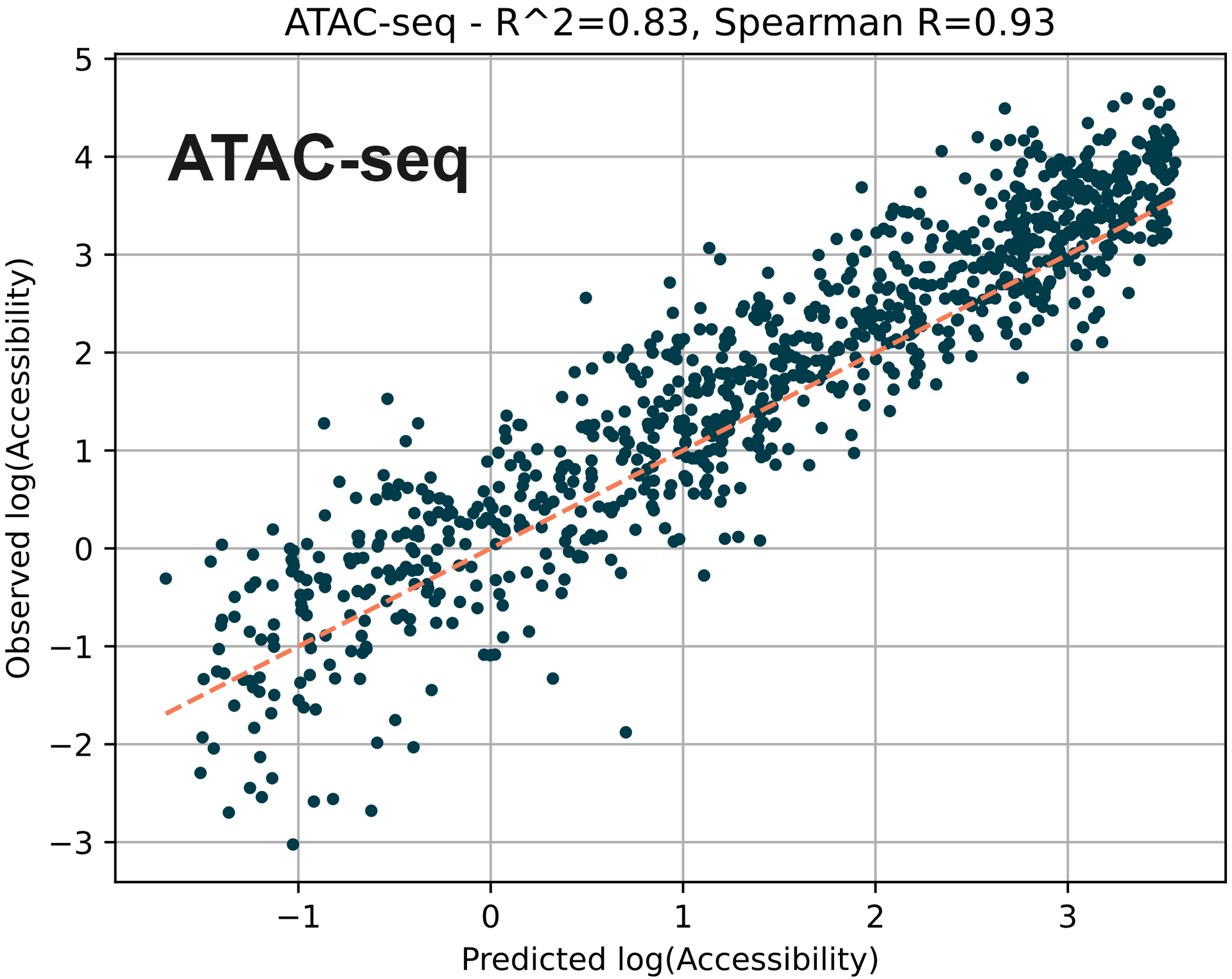
Using machine learning to predict chromatin accessibility around TSS using measured 5hmC and 5mC. ATAC-seq is often used to indicate open chromatin owing to the ability of the transposase enzyme to tagment accessible regions of the genome.
Here, we measured mean 5mC and 5hmC fractions for 1kb regions around each TSS (transcriptional start site) and used them as features. Using each of these features, along with the number of CpGs and length of the region, we trained a simple regression model (XGBoost) on publicly available ATAC-seq data for the E14 mESC cell-line.
The training set used all chromosomes except chromosome 8. Plotted are predicted versus observed ATAC-seq chromatin accessibility values for the test dataset (chromosome 8).
Here we find that the accessibility predicted by the model correlates with accessibility on chromosome 8 with an R2 of 0.83.
Enhancer states:
- Repressed: does not enhance gene expression
- Primed: is ready to activate gene expression
- Active: actively enhances gene expression
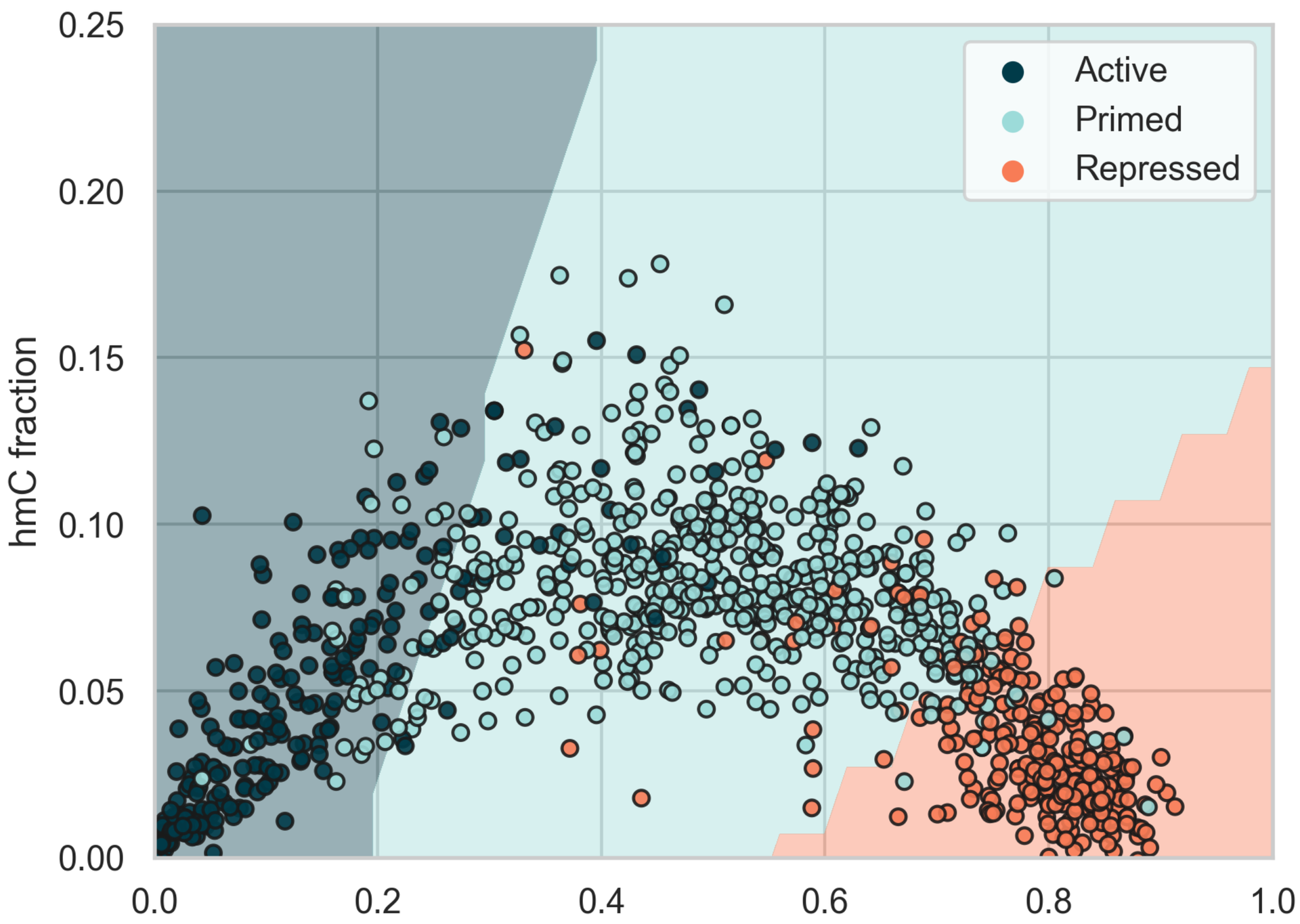
Classification of enhancer states in the 5mC vs. 5hmC fraction space.
Enhancers are cis-acting regulatory regions that have a profound effect on cell-type specific gene expression programs. These regions are typically classified by histone modifications in flanking nucleosomes.
- Active enhancers (H3K4me1 & H3K27ac) have low 5mC and 5hmC levels
- Primed enhancers (H3K4me1 but not H3K27ac) moderate 5mC and high 5hmC levels
- Repressed enhancers (H3K9me3) have high 5mC and low 5hmC levels
An SVM model is able to classify the three groups with 85.5% accuracy based on their 5mC and 5hmC levels only (different shades represent the decision boundaries of the classifier).
Here we present duet multiomics solution evoC, an enzymatic method that reads the four canonical bases in DNA together with the power to read epigenetic information encoded in DNA. This information is encoded in two important cytosine modifications, 5mC and 5hmC, that appear to have an opposing pattern when examined through the lens of gene expression. 5mC appears to be repressive – it correlates with silenced regions of the genome. In contrast, high 5hmC levels are thought to be found in regions of the genome that appears to be active.
Here, we examine patterns of variation in these two cytosine modifications resolved for the first time in a single workflow in mouse embryonic stems cells. We used these measurements to train models that can be used to predict accessible chromatin (TSS), RNA-sequencing both in bulk and newly synthesised RNA. Finally, we show that enhancer classification in this cell-line, which are often important for establishing tissue specific gene expression programs, can be grouped using cytosine modifications in DNA. This demonstrates the power of reading all six bases as a new lens to examine the dynamic information encoded in DNA.
- Simultaneous sequencing of genetic and epigenetic bases in DNA, Füllgrabe and Gosal et al., Nature Biotechnology (2023). (duet multiomics solution technology paper)
- Acute depletion of the ARID1A subunit of SWI/SNF complexes reveals distinct pathways for activation and repression of transcription. Blümli S, et al. Cell Rep (2021).
- Cell Transcriptomics CRISPR-Activation Screen Identifies Epigenetic Regulators of the Zygotic Genome Activation Program. Alda-Catalinas C, et al. Cell Syst. (2020).
- Pioneer activity distinguishes activating from non-activating SOX2 binding sites. Maresca M, et al. EMBO J (2023).
- PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cruz-Molina S, et al. Cell Stem Cell (2017).