- Jean Teyssandier
- Nicholas Harding
- Sabri Jamal
- Michael J. Wilson
- Gary Frewin
- Nicola Wong
- William Stark
- Mark S. Hill
- Páidí Creed
We present a computational toolkit to analyse 5mC and 5hmC modifications at scale and describe its performance on a novel liquid biopsy dataset.
Methylation data has diverse applications in cancer, including early-stage diagnosis through liquid biopsy, classification to guide treatment pathways, and prognosis. However, analyzing methylation data poses significant challenges, as it is constrained by scalability and usability issues.
Our recently introduced technology, duet multiomics solution evoC, enables the reading of 6-base information (A, T, G, C, 5-mC and 5-hmC) from DNA, further amplifying the complexity and scale of datasets generated in a single sequencing experiment. To address this, we present a fast and scalable array-based python package (called modality) for the analysis of 6-base genomes, using multi-core out-of-memory processing to enable extremely efficient computation, even for datasets that are too large to fit into memory.
Unlike existing tools that exceed typical laptop memory with ~10 samples, the modality package can efficiently analyse (e.g. DMR calling) a colorectal cancer liquid biopsy dataset of over 100 samples in minutes on a standard laptop.
modality combines efficient computation with tools for exploratory (e.g., plotting, methylation summaries) and downstream analyses (e.g., DMR identification, PCA). Designed for efficiency and ease of use, it enables users to rapidly transition from raw data to actionable insights and publication-ready results. As multiomic data become the standard in cancer research, our data structure supports the integration of additional data types, allowing us to handle combined genomic and epigenomic data from solutions like duet evoC. This will enable streamlined and efficient multiomic analysis to uncover deeper biological insights.
(b)
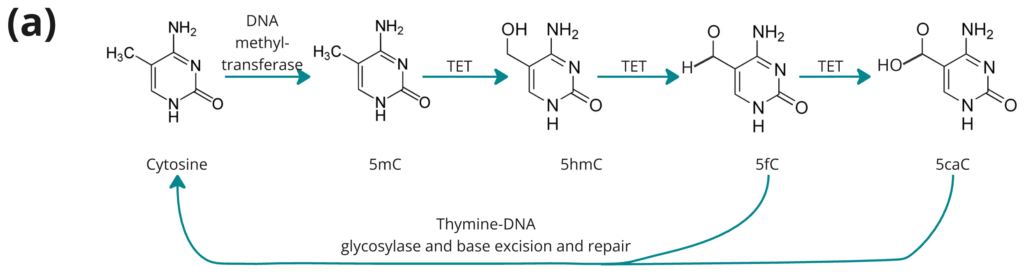
- TET-mediated demethylation pathway
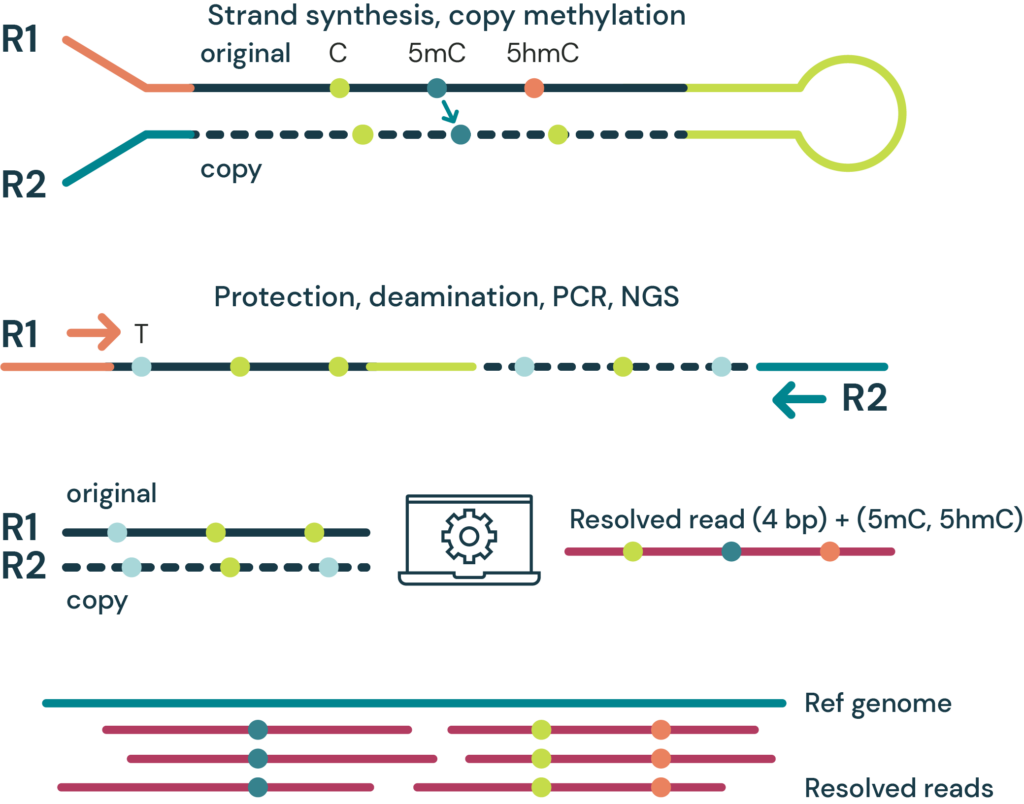
- duet multiomics solution evoC – a 6-base sequencing technology that reads all four canonical bases plus 5mC and 5hmC¹ via strand copy, 5mC copy and 5mC + 5hmC protection enzymatic steps.
- The duet multiomics solution evoC works as an end-to-end solution comprising reagents & bioinformatics pipeline
(c)
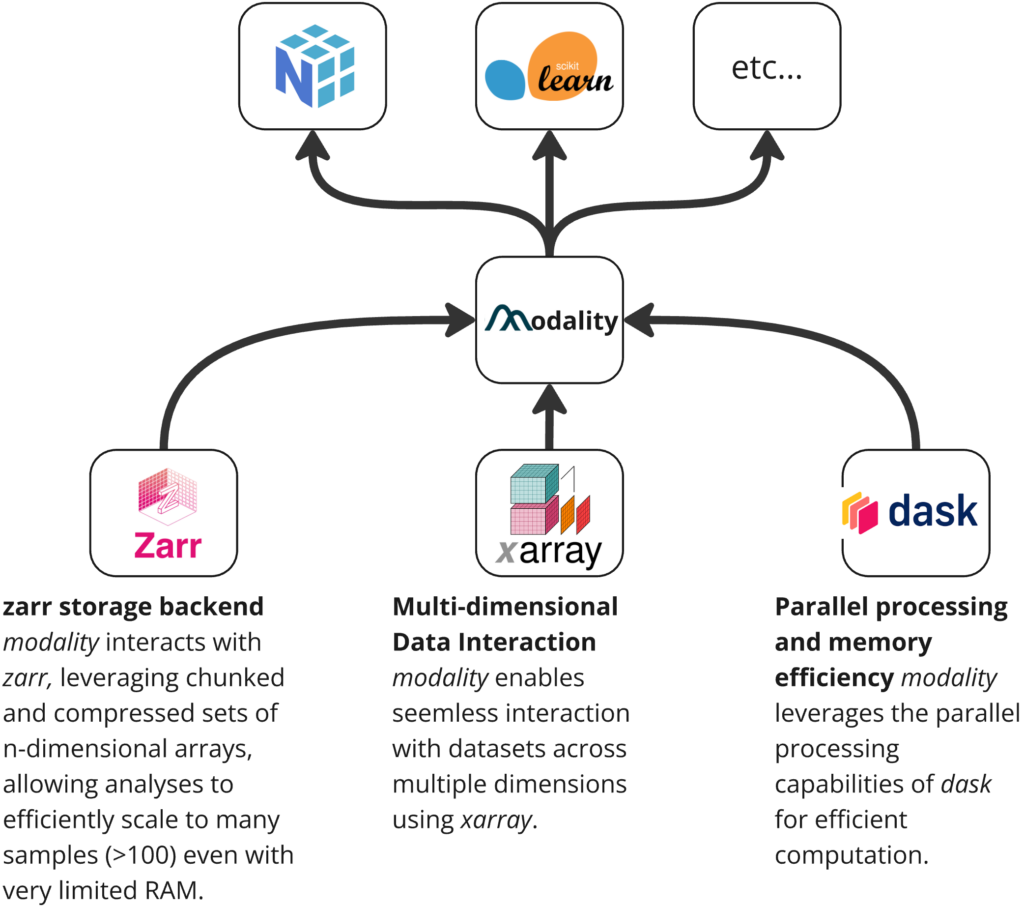
modality is built around three core python packages: zarr, xarray, and dask. These are powerful modern data science packages which collectively allow modality to deal with larger-than-memory data arrays in an expressive and paralelised fashion. This foundation means that analyses that would previously require long run times and extensive compute infrastructure can now run quickly on one’s laptop – speeding up iterative data analysis.
The core data structure used by modality is the ContigDataset. This contains arrays that represent methylation counts as well as accompanying arrays which encode the coordinates. This object also provides a set of easy-to-use and efficient methods for working with them. Each of the arrays are chunked Dask arrays allowing extremely efficient computation.
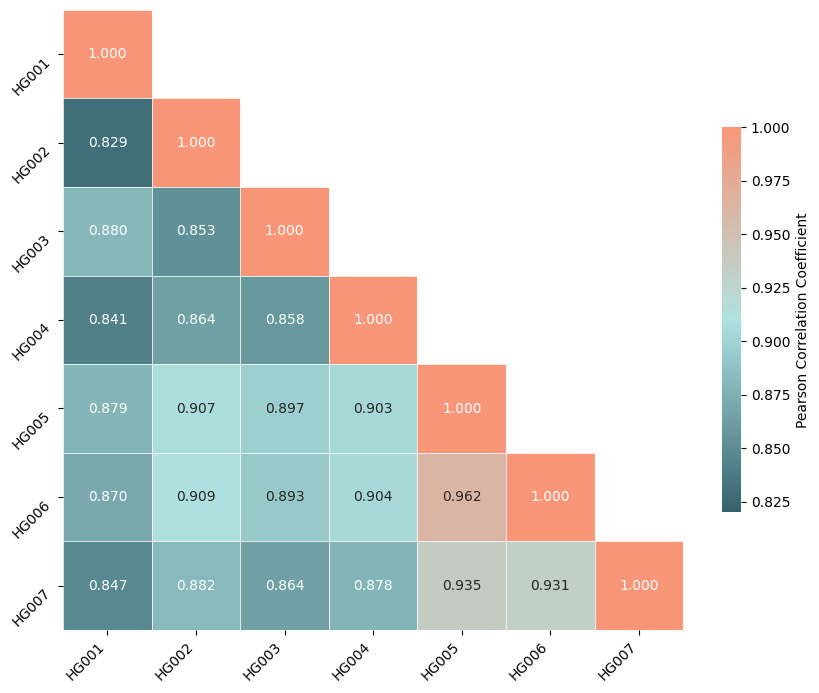
Pearson correlation coefficients for the 7 samples in the GIAB data
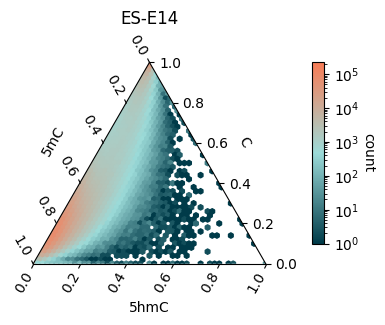
Ternary plot of C, 5mC, and 5hmC in a mouse ES-E14 sample
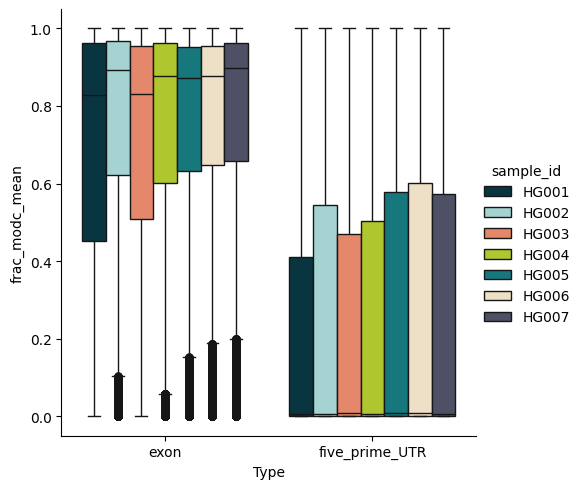
Distribution of 5mC fraction in exons and 5’UTRs in the GIAB data using modality’s feature extraction capabilities
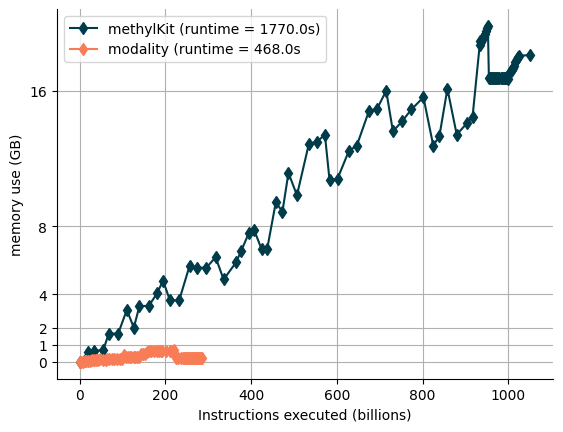
Benchmarking of modality vs. MethylKit on DMR calling
We conducted a performance comparison between the DMR caller in modality and the one from MethylKit. A key limitation of MethylKit is its high memory consumption when reading text files, which eventually leads to it exhausting the available system memory. In contrast, modality is memory-efficient, allowing for genome-wide DMR calling on a standard laptop.
To evaluate real-world usability, we tested both tools on a colorectal cancer (CRC) dataset consisting of 58 samples divided into two conditions (control vs stage I). Using modality, we were able to perform genome-wide DMR calling in just 12 minutes when tiling the genome into 2kb windows. In comparison, MethylKit was unable to process the dataset in a single run, as it ran out of memory while attempting to read the input files. This highlights modality’s scalability and efficiency, making it a more practical choice for large-scale epigenomic analyses.
modality can process large cohorts of samples genome-wide on a standard laptop. Here we show an example of an analysis on a colorectal cancer dataset.
- We call DMRs between Control and Stage I patients on promoters (defined as 1kb upstream of TSS) of protein-coding genes from the GENCODE hg38 annotation database.
- We identify significant DMRs by filtering by q-value and mean methylation difference between the two groups.
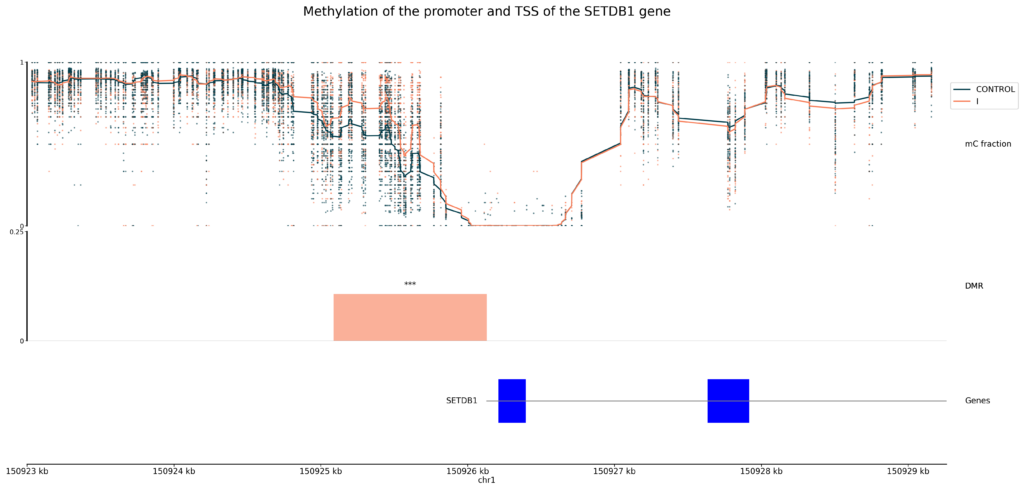
- Below we show an example of a differentially methylated promoter upstream of the SETDB1 gene, which is known to promote CRC progression [2], using modality’s genomic tracks plot feature.
To address the difficulties of analysing methylation data, we present modality, an efficient and scalable analysis package for 5- and 6-base genomes.
- The package is built on a core set of performant data science libraries and roots the user into a powerful ecosystem for data analysis in Python.
- modality has an intuitive API providing powerful analysis and visualisation methods, enabling users to gain insight from duet multiomics solution evoC directly on a laptop.
- Moving forward, the underlying data structure used by modality is very extensible, allowing other data modalities to be incorporated and analysed alongside the methylation data modalities shown here.


