Accelerate your journey from multiomic data to biological discovery with modality XPLR – a high‑performance, low‑code analysis software for analysing 6‑base genome data.
Move from raw sequencing counts to insight with intuitive differential methylation analysis, biological QC, and feature extraction—all in a single, scalable workflow, on a standard laptop.
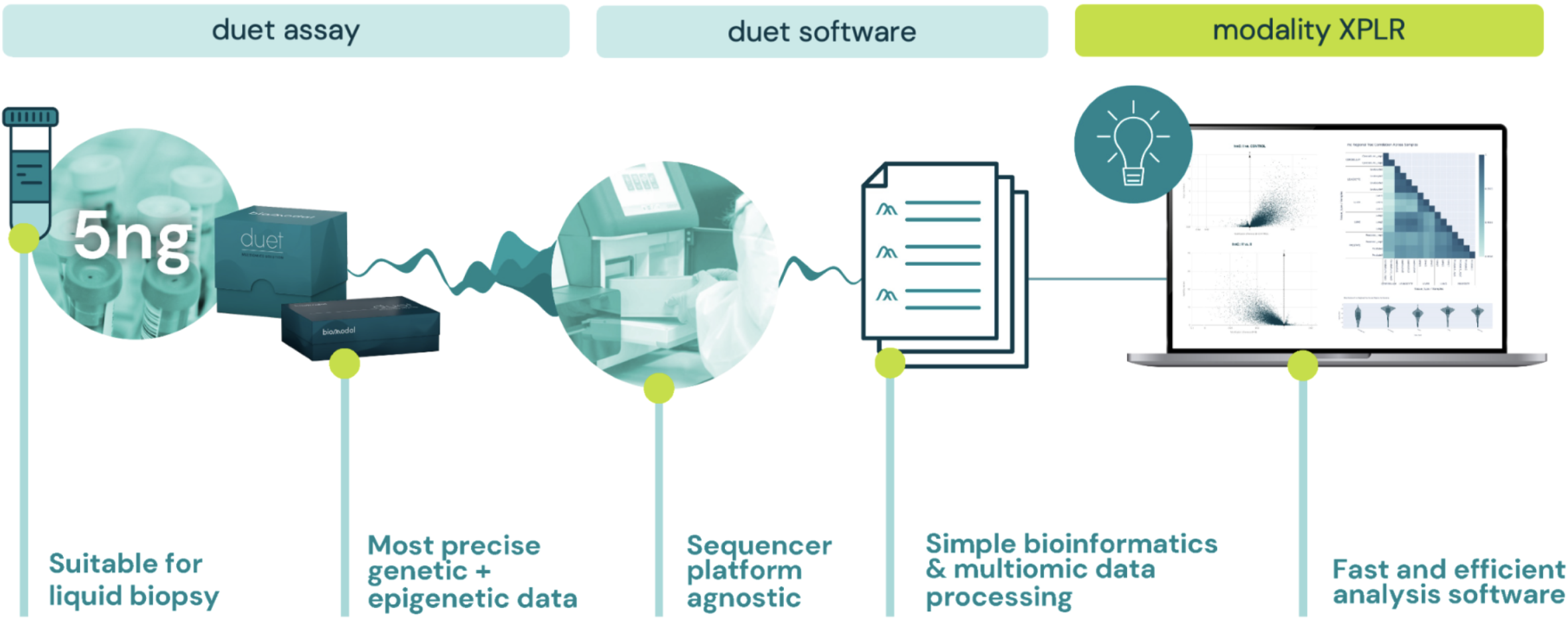
modality XPLR is an accessible and scalable software tool designed for scientists and translational researchers to uncover disease-relevant epigenetic signatures, link methylation to gene regulation, and support biomarker discovery—empowering you to identify, classify, and monitor disease.
With a low-code, command-line interface and comprehensive documentation, modality XPLR provides multiomic analyses and advanced publication-ready visualisations.
With modality XPLR, you can go further than basic sequencing checks to understand your datasets in biological context. Biological QC tools empower you to identify patterns and relationships in multiomic data, using intuitive correlation matrices and PCA plots. Instantly visualise sample similarities, spot outliers, differentiate subtypes and reveal underlying biological signals—so you can explore, interpret, and act on your results with confidence.
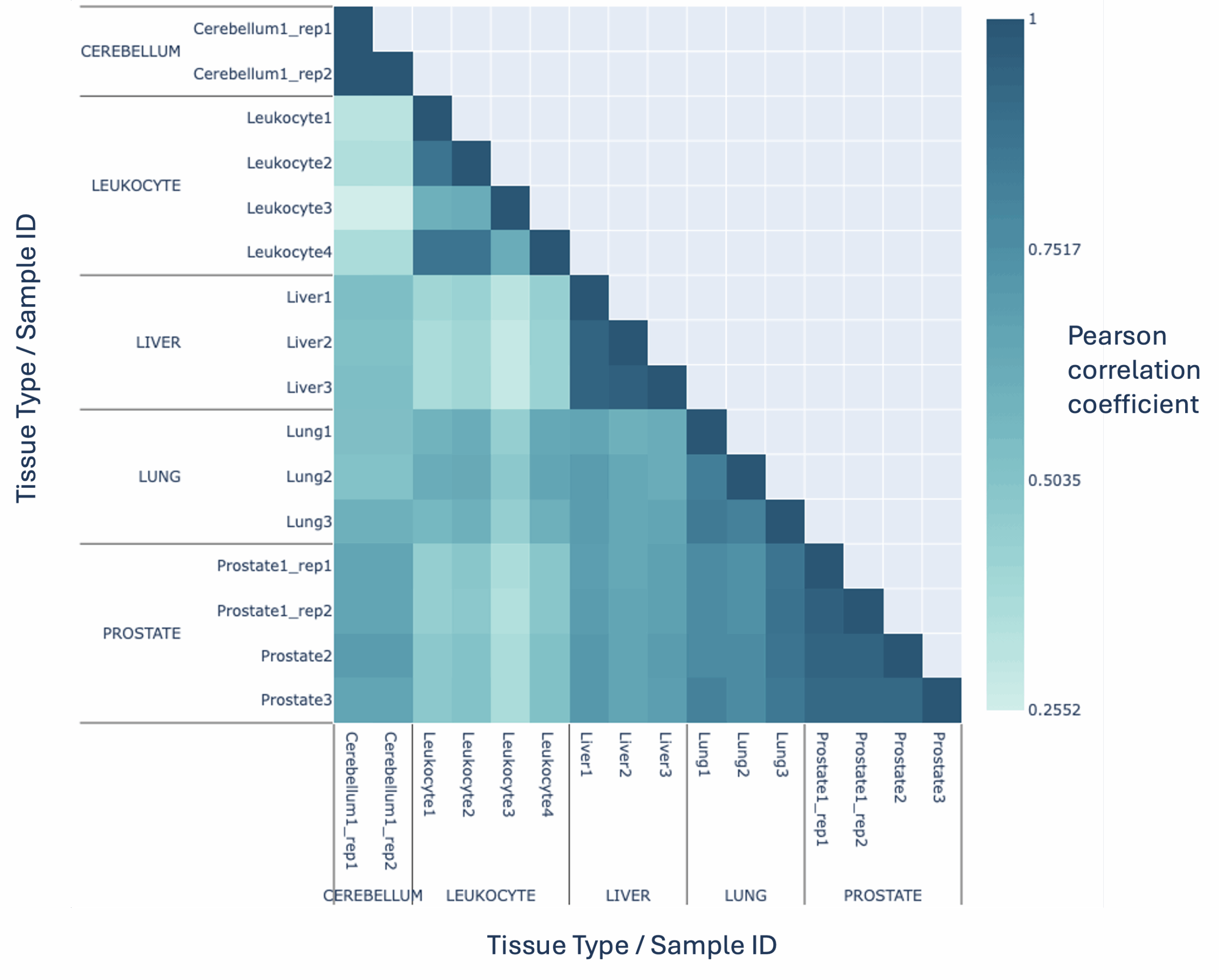
Correlation of hmC fractions across gene bodies (hg38), between tissue samples. These plots can be used to observe sample relationships for 5mC and 5hmC, and are included in the modality XPLR Biological QC report as standard.
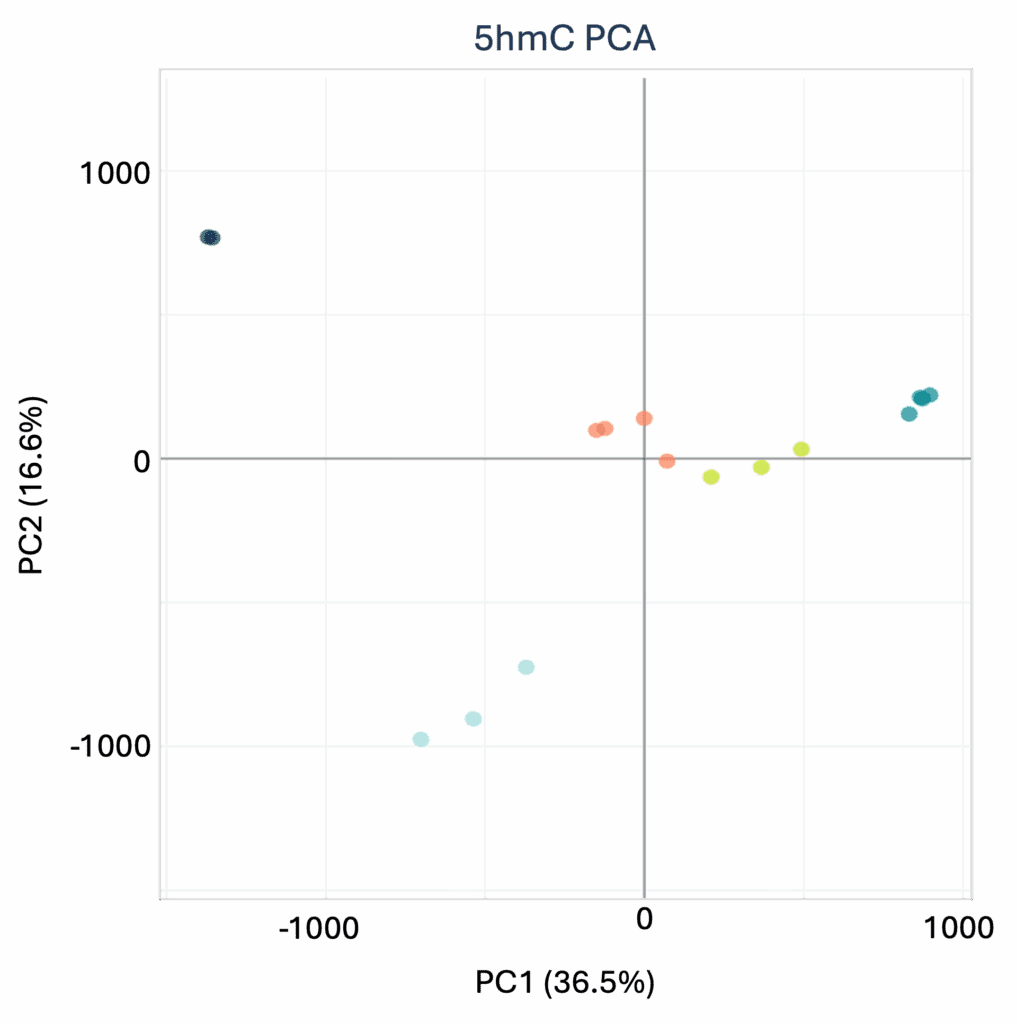
Principal Component Analysis for 5mC between tissue samples. These plots can be used to observe sample relationships for 5mC and 5hmC, and are included in the modality XPLR Biological QC report as standard.
modality XPLR makes it easy to identify differentially methylated regions (DMRs) across the genome. Flexible region definitions via BED file input, robust statistical testing, and advanced correction methods help you pinpoint significant methylation differences between groups—revealing disease signatures, treatment responses, and better biomarkers.
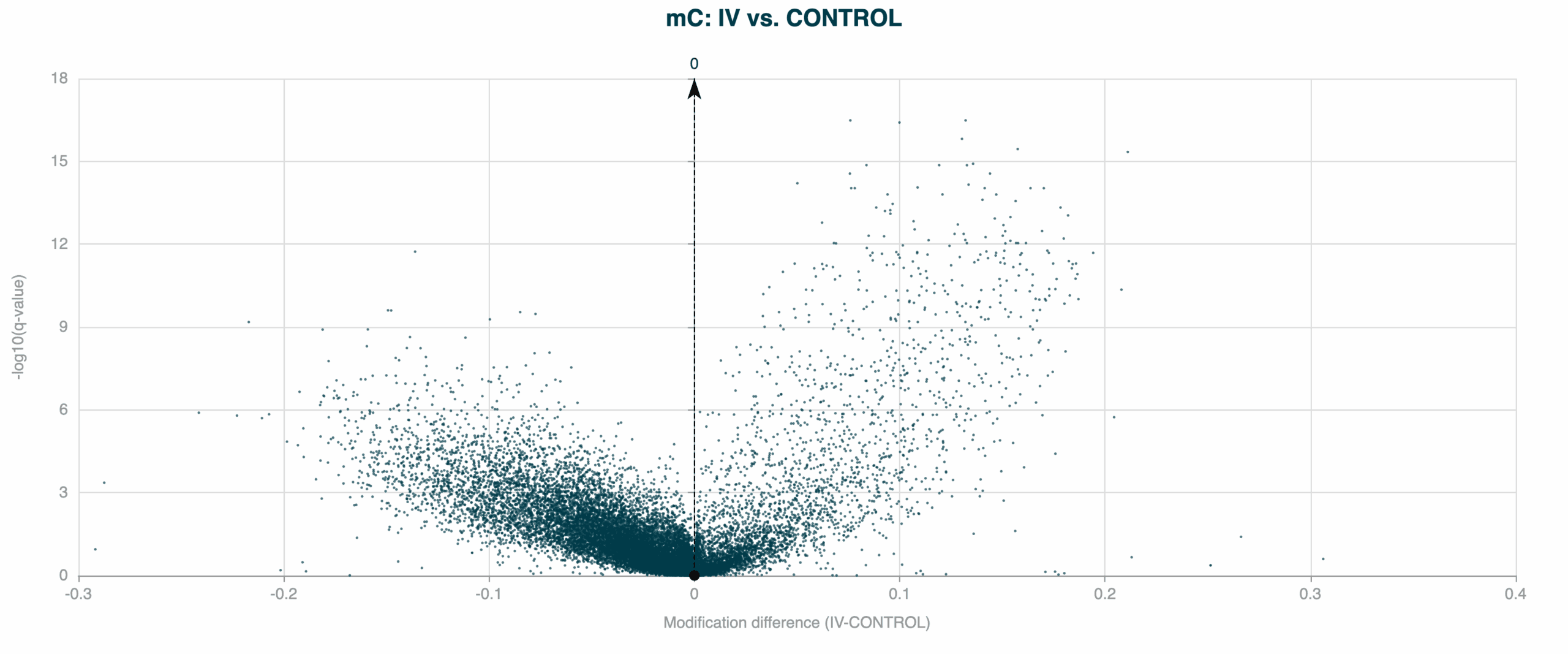
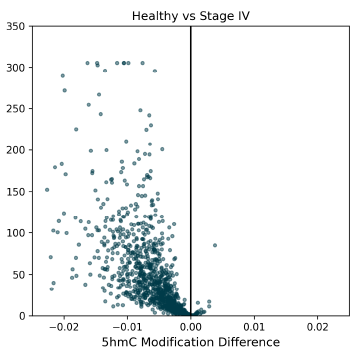
5mC DMRs between Healthy Control and Stage IV CRC cfDNA samples, showing a high density of 5mC hypomethylation (negative modification difference when compared to controls), which is typically associated with gene repression.
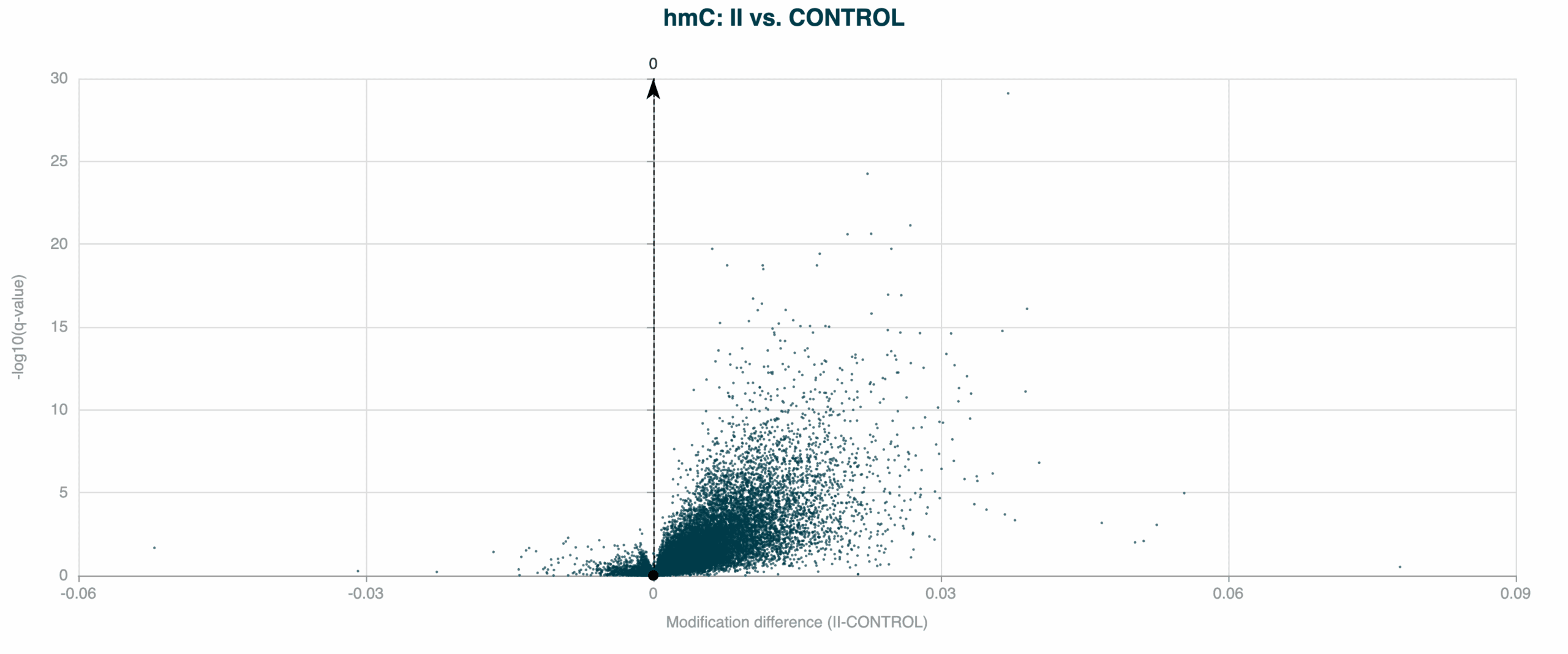
5hmC DMRs between Healthy Control and Stage II CRC cfDNA samples, showing a trend early-stage 5hmC hypomethylation. 5hmC is an intermediate mark on the TET-mediated demethylation pathway, and is indicative of later gene activation.
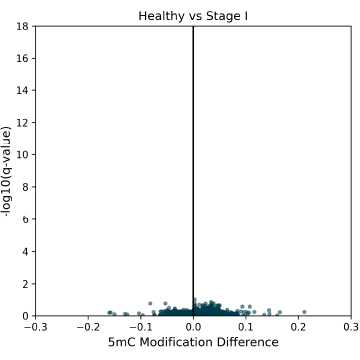
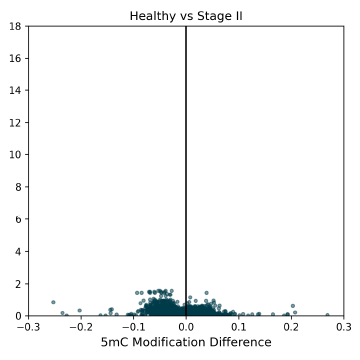
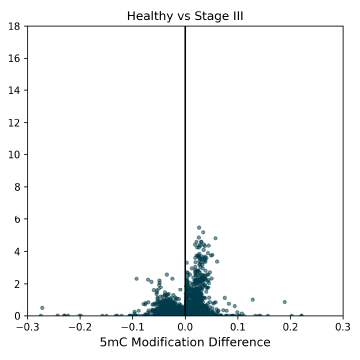
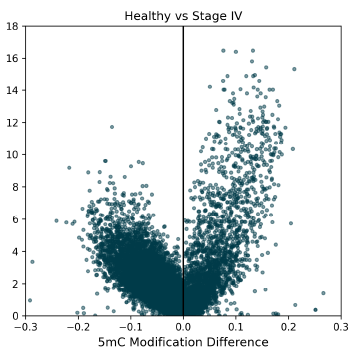
5mC DMRs for promoter regions (1000bp upstream of transcription start sites) between Healthy Control and CRC cfDNA from early to late-stage disease. Significant 5mC DMRs are most detectable in late stages of disease progression. Overdispersion correction applied.
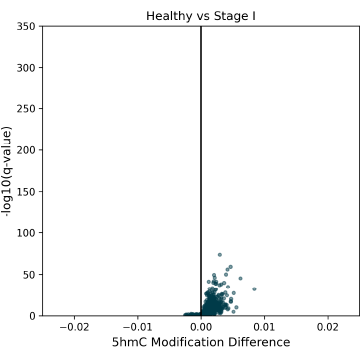
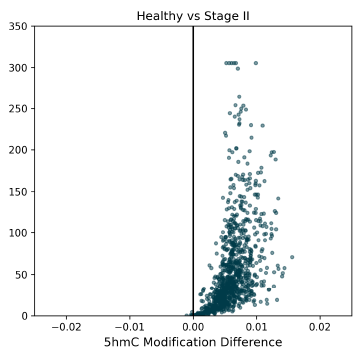
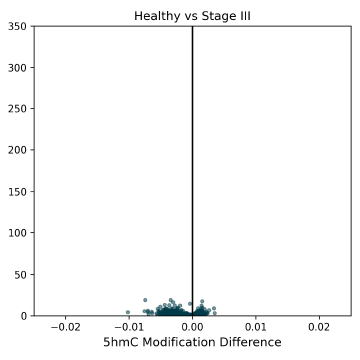
5hmC DMRs for HCT116 super-enhancer regions between Healthy Control and CRC cfDNA from early to late-stage disease. Significant hypermethylated 5hmC DMRs are detectable at early stages, and help to explain the late-stage hypomethylation in 5hmC (this figure) and 5mC (Figure 5).
Move beyond summary statistics and visualise methylation changes in biological context, directly in gene track plots. Explore DMRs alongside genes, promoters, and regulatory elements—making it easy to interpret results, pinpoint biologically relevant changes, and understand the role of 5mC and 5hmC in genome dynamics. Instantly connect your findings to functional genomics and accelerate biological discovery.
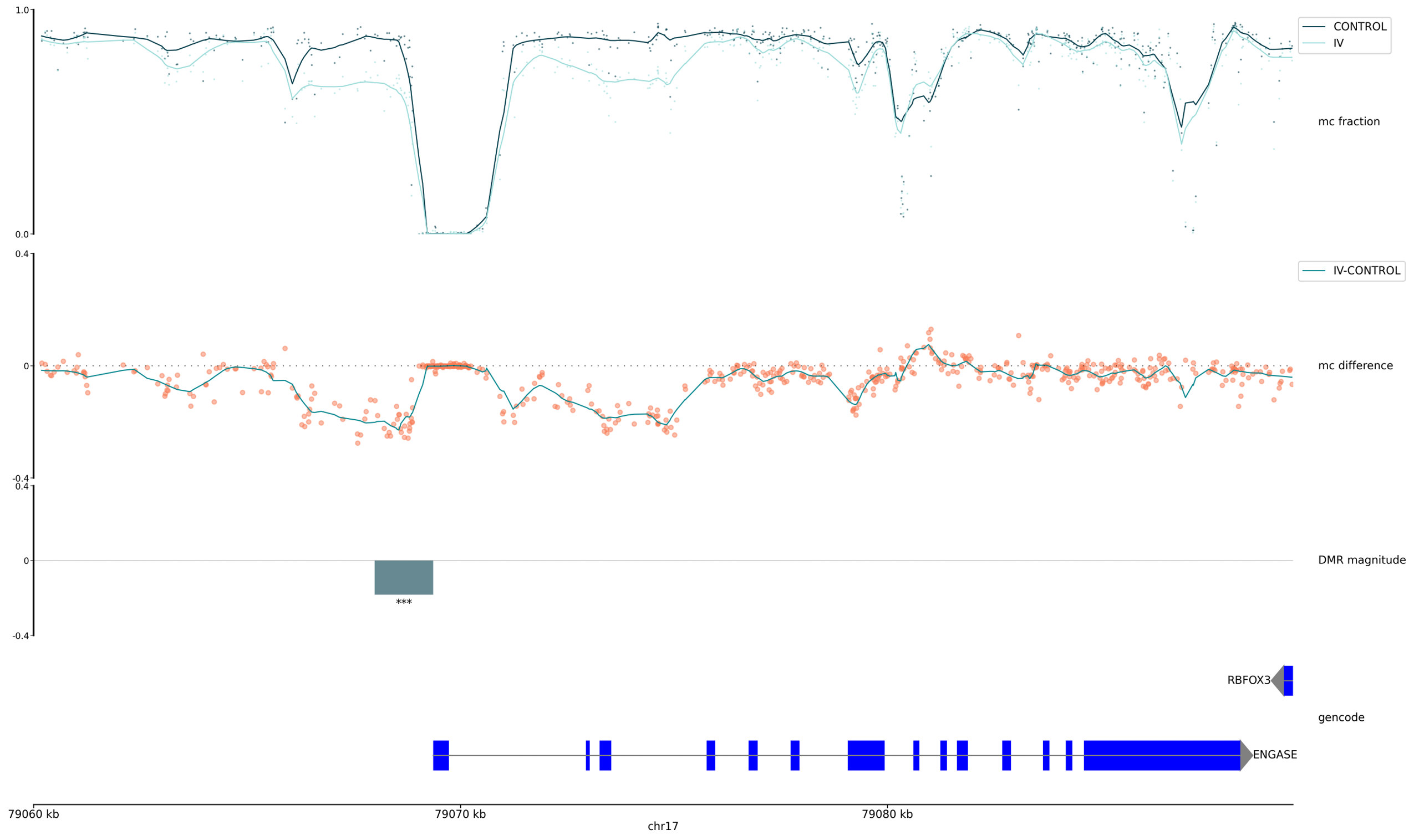
Methylation fraction by genomic position (points) and smoothed (line) at CpG contexts for Control (dark teal) and Test (light teal) groups.
Methylation difference by genomic position (points) and smoothed (line) at CpG contexts between the two groups. Positive values indicate hypermethylation and negative values indicate hypomethylation relative to the Control group.
Magnitude of modification difference for defined DMR regions (block), Positive values indicate hypermethylation and negative values indicate hypomethylation relative to the Control group. Significance level indicated by the number of asterisks.
Annotation of genomic features via user-supplied GTF/GFF reference, showing genes and exons (blue boxes).
Track plot for the ENGASE gene, showing a hypomethylated 5mC DMR between Healthy Control and Stage IV CRC cfDNA. The four tracks are methylation fraction, methylation difference, DMR magnitude and genome annotation. The DMR significance (p-value) is indicated by the number of asterisks (≤0.001), the difference between 5mC fractions is highlighted by divergence of the mean methylation between the two groups (red arrow), immediately prior to the ENGASE TSS (annotation track).
Efficiently subset data, build feature sets, visualise and export methylation counts in community-supported file types (CX Report, bedmethyl, bedgraph and Bismark). The modality XPLR feature extraction tools support region-based exploration of methylation statistics, by genome annotation or DMR result. Feature extraction is a valuable precursor to downstream classifier development.
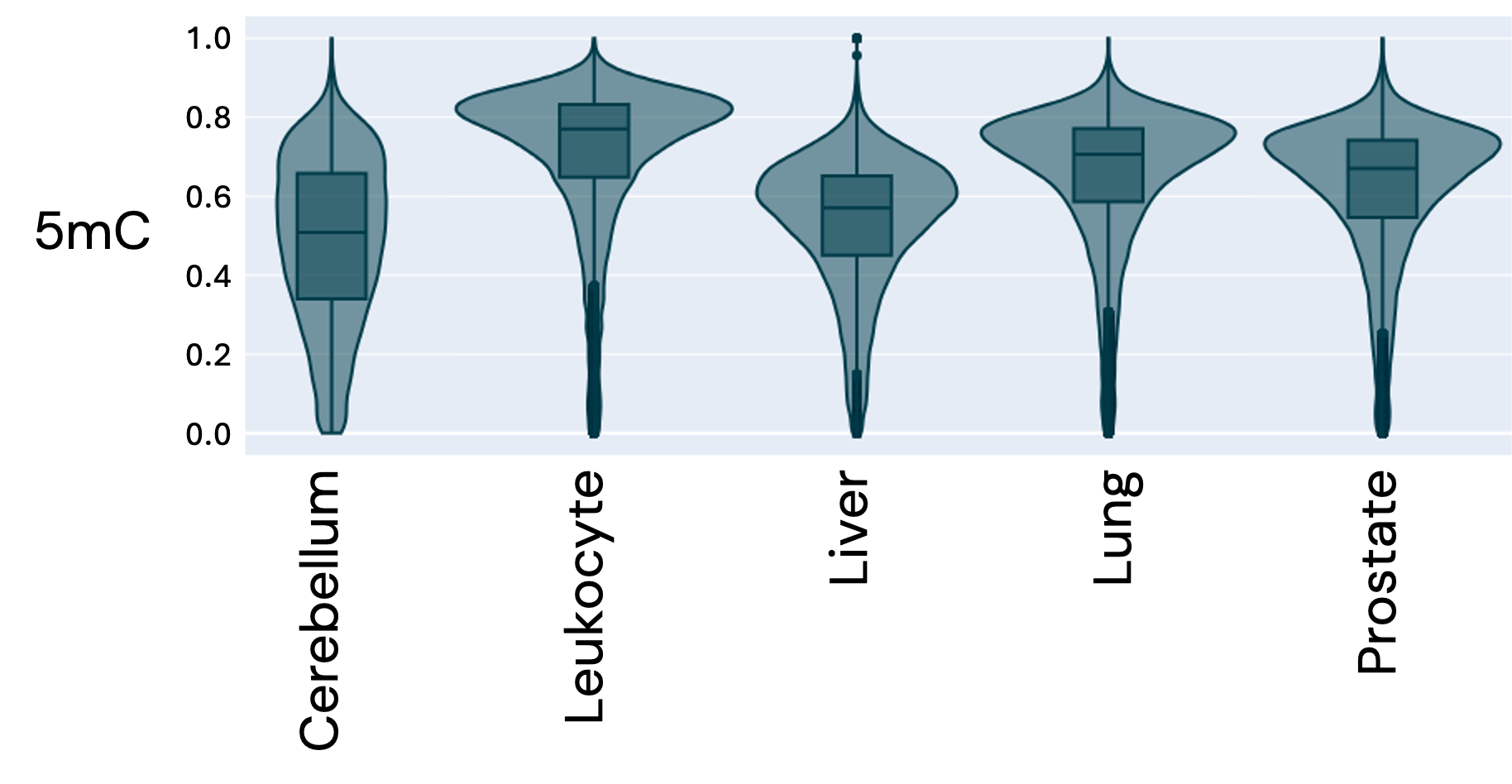
Violin plot showing the distribution of 5mC fractions over gene bodies by tissue type, indicating tissue-specific differences in methylation profiles
We’ve validated modality XPLR DMR calling across a range of analytical parameters, using a truth set of simulated DMRs spiked into real cfDNA data. The effect of region size, CpG coverage and sample size are shown with respect to DMR sensitivity and false discovery (Figure 9). Next, using dynamic, genome-wide pre-segmentation method guided by the distribution of CpGs, we demonstrate the DMR sensitivity and false discovery rate according to DMR effect size (Figure 10).
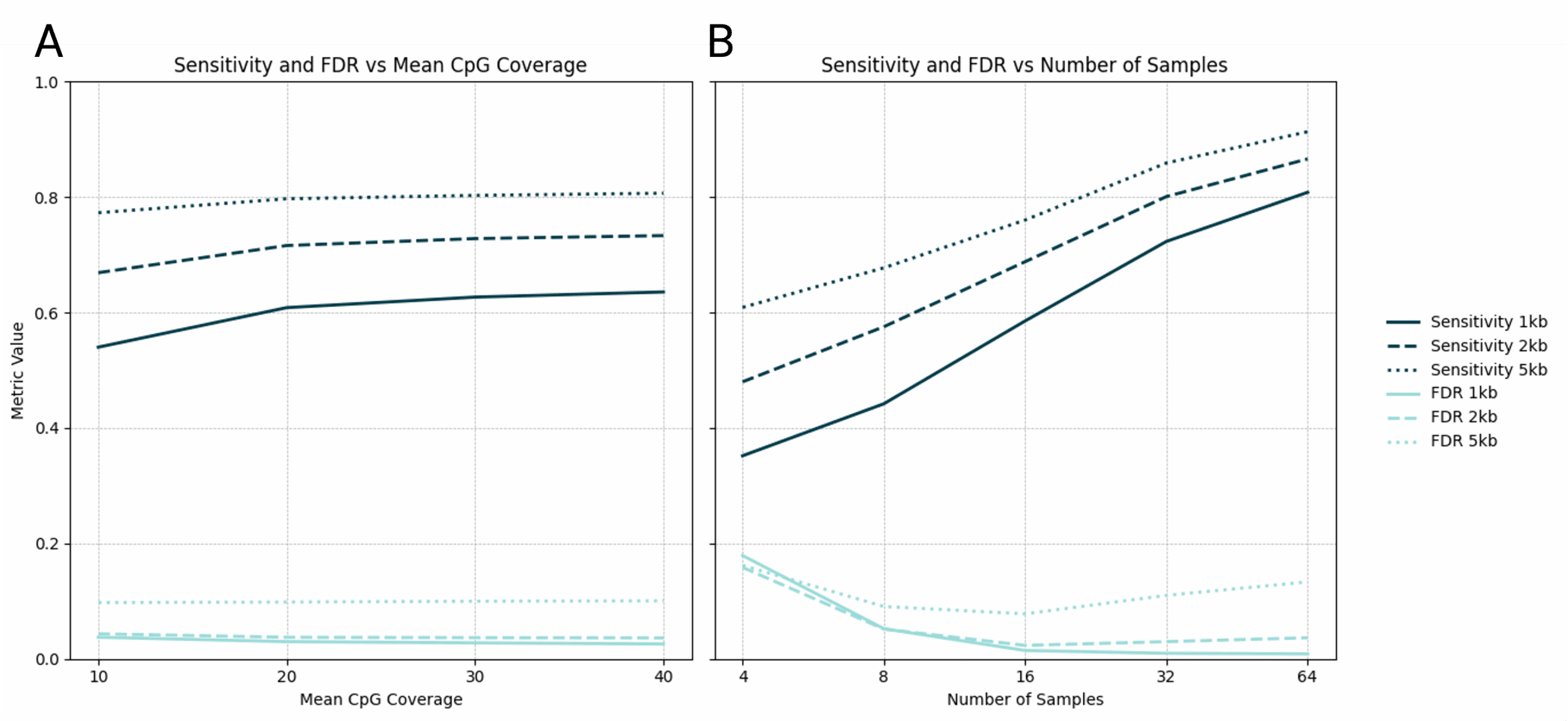
modality XPLR 5mC DMR calling sensitivity and FDR, for region sizes of 1Kb, 2Kb, and 5Kb, by (A) mean strand-merged CpG coverage with 8 samples per group and (B) sample size with up to 32 samples per group (64 total) and a mean strand-merged CpG coverage of 21x.
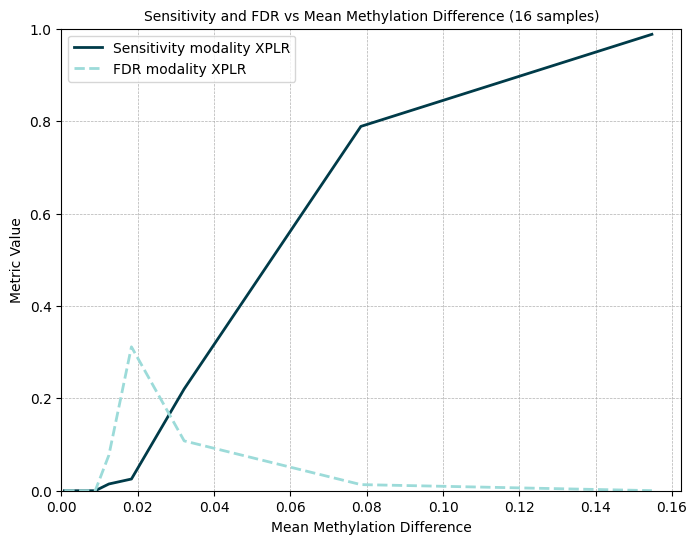
Sensitivity and FDR for 5mC DMR calling for pre-segmented regions by mean methylation difference (effect size).
To assess real-world usability, we tested modality XPLR and methlyKit (an alternate community tool) using an 8-sample pilot dataset on a. standard laptop. Using modality XPLR, DMR calling over 19,382 promoter regions completed in under 7 minutes. However, methylKit failed to load the input files due to memory constraints.
This comparison highlights modality XPLR’s scalability and efficiency, making it a practical solution for large-scale epigenomic studies.
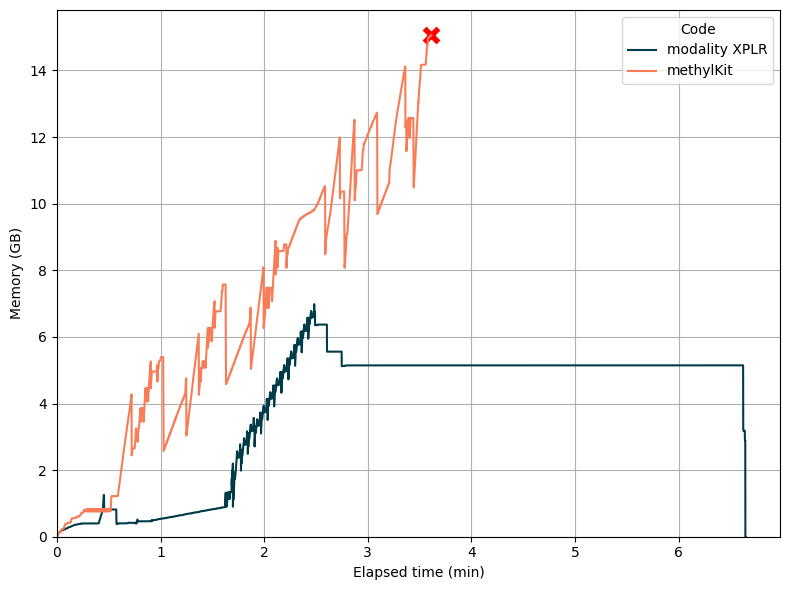
Memory usage over time for genome-wide DMR calling on 19,382 promoters with 8 samples on a standard laptop (4 cores, 16 GB RAM). Whilst modality XPLR efficiently completes the analysis in a round 7 minutes, methylKit cannot complete the operation due to memory exhaustion.