We are excited to participate in the Genomics symposium hosted by the NIEHS Molecular Genomics Core laboratory.
NIEHS
Building 101, Rodbell Auditorium
111 TW Alexander Drive
RTP, N. Carolina
Visit biomodal at booth
We are excited to participate in the Genomics symposium hosted by the NIEHS Molecular Genomics Core laboratory.
NIEHS
Building 101, Rodbell Auditorium
111 TW Alexander Drive
RTP, N. Carolina

Field Application Scientist
biomodal
The predominant cytosine modification in DNA is methylcytosine (5mC). This base appears to exert a profound impact on gene expression patterns and is commonly linked to transcriptional repression. The introduction of hydroxymethylation (5hmC) adds a new layer of complexity to the conventional comprehension of DNA methylation dynamics, which is often thought to correlate with gene expression. Constrained to measuring four states of information, existing NGS-based technologies sacrifice genetic information for modification calling without distinguishing between these two important modification states. Genetic and methylation data combined together presents a unique opportunity to measure these cytosine modifications against gene expression.
duet multiomics solution evoC is a new sequencing technology that resolves all four genetic bases alongside the ability to distinguish the modification status of cytosines, discriminating 5mC from 5hmC (6 base calling). The technology consists of pre-sequencing library prep with enzymatic conversion of DNA together with an analysis pipeline, achieving base resolution of genetics and epigenetics at high accuracy. Here we present the method, alongside a high-depth sequencing analysis of mouse embryonic stem cells. We demonstrate the potential of epigenetic modifications encoded in DNA to predict other important potential modulators of gene expression such as open chromatin and enhancer states as well as RNA expression itself.
Here are the relevant biomodal resources for information. Find poster presentation information, case studies, interviews, and more.
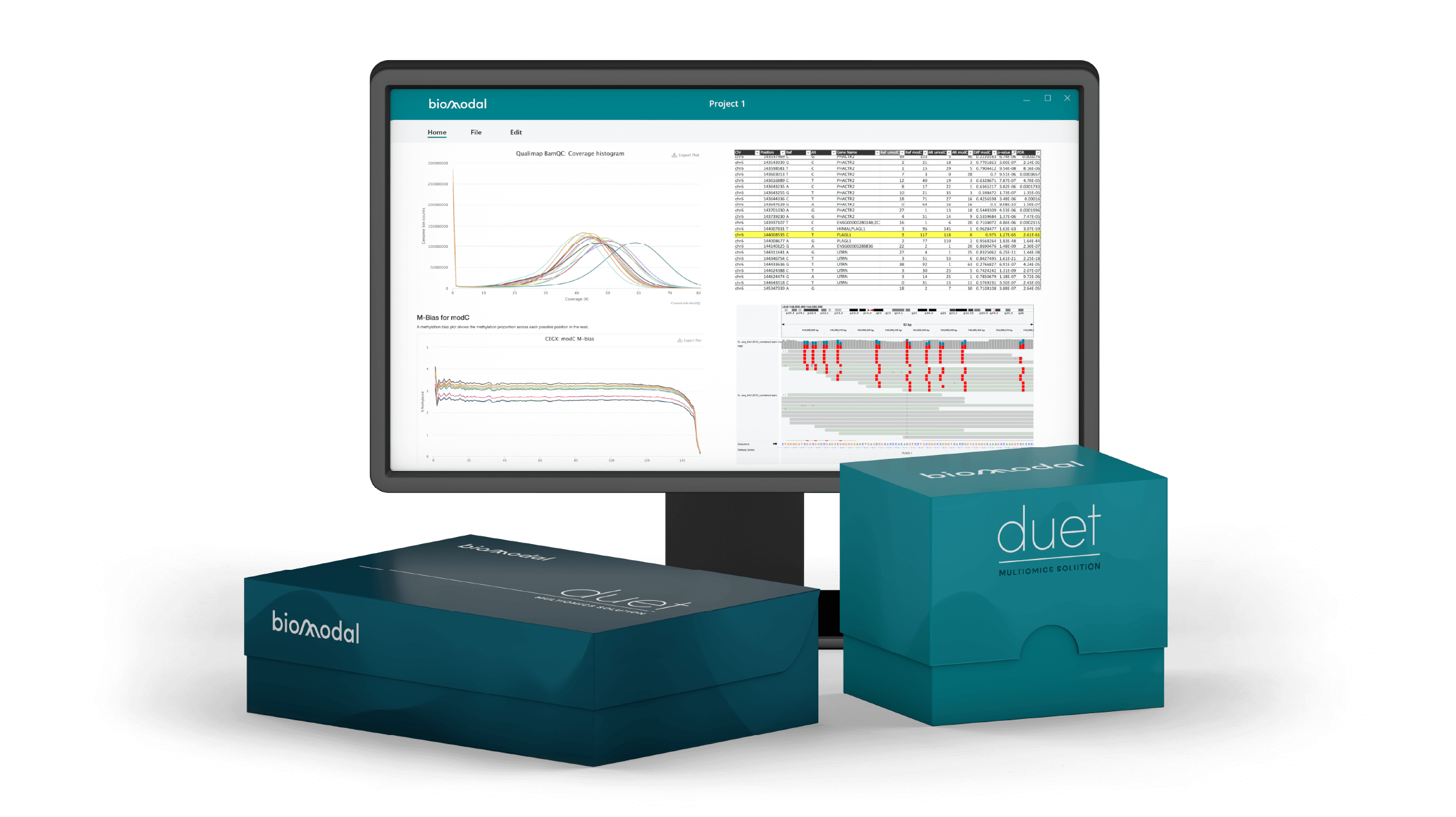
Unlock the power of the 6-base genome with duet evoC data, providing unprecedented insight, including A, C, G, T plus 5mC and 5hmC, in a single read. Learn more
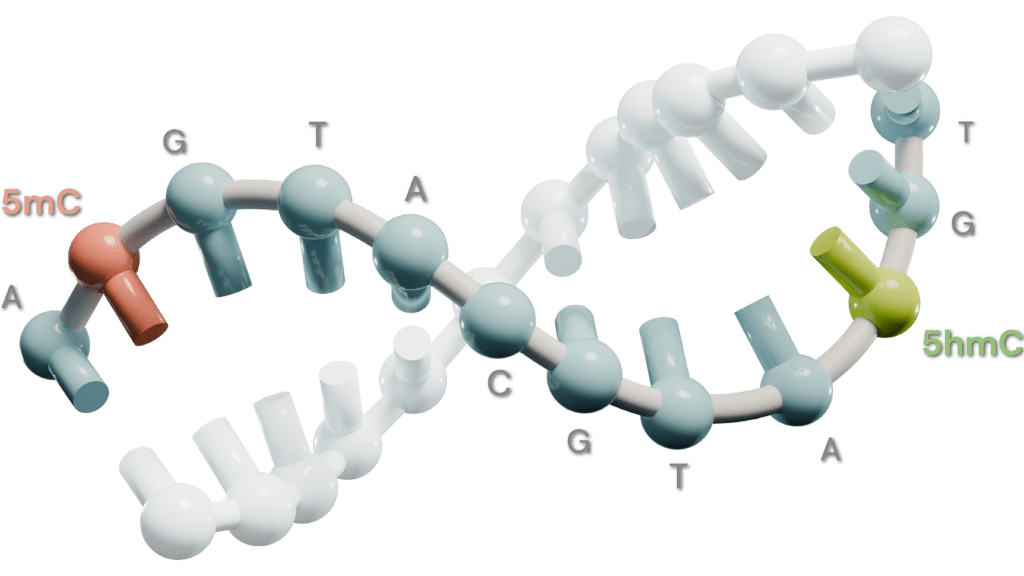
The 6-base genome expands our understanding of genetics by incorporating both genetic & epigenetic data to get a clearer picture of health and disease biology.
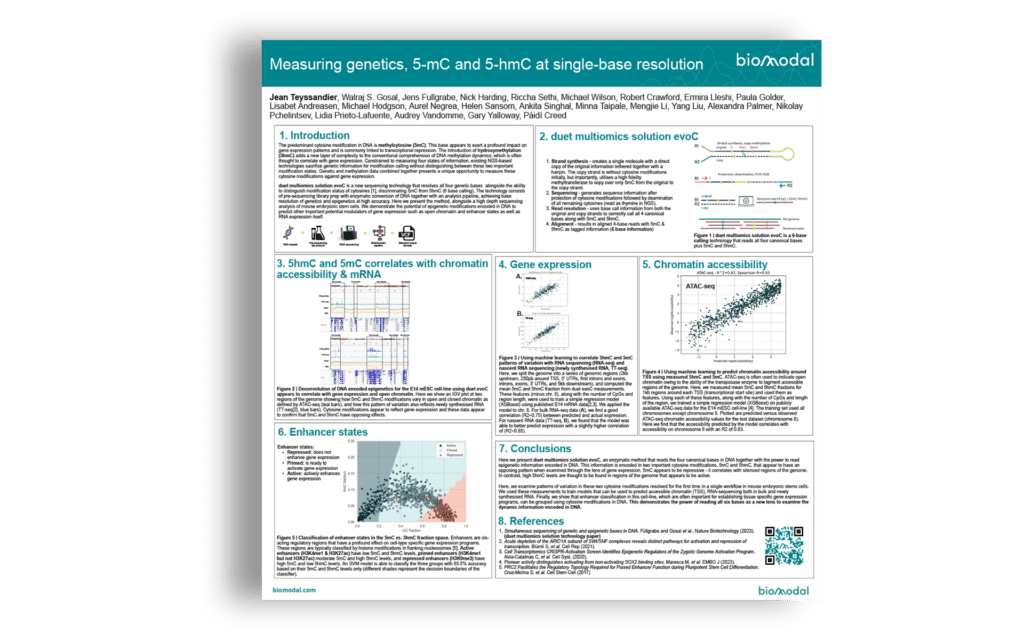
Using the 6‑base genome, genetics, 5mC and 5hmC, to gain multimodal insights into genome organisation and gene regulation

Generating multiomic data from as little as 5ng of cfDNA that can be used to identify SNPs, CNVs, differentially methylated regions (DMRs), and variant associated methylation (VAM). Using these techniques, methods to identify new potential biomarkers and fragment length profiles are also discussed.
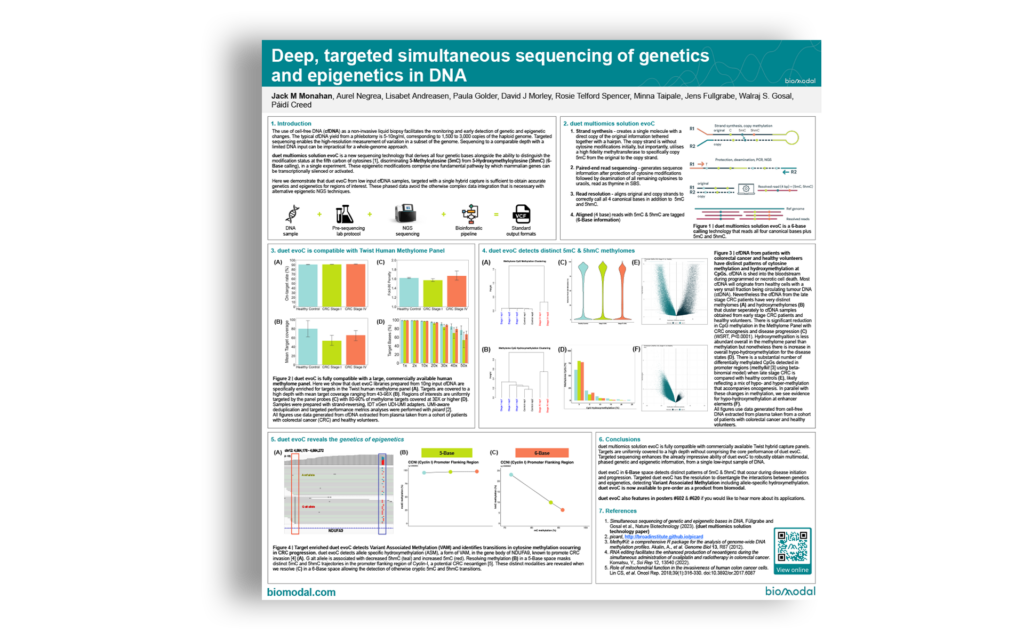
DNA comprises molecular information stored in genetic and epigenetic bases, both of which are vital to our understanding of biology. The interplay between genetics and the DNA epigenome orchestrates complex biological phenomena as diverse as cell fate, ageing, the response to environmental stimuli, and disease pathogenesis.