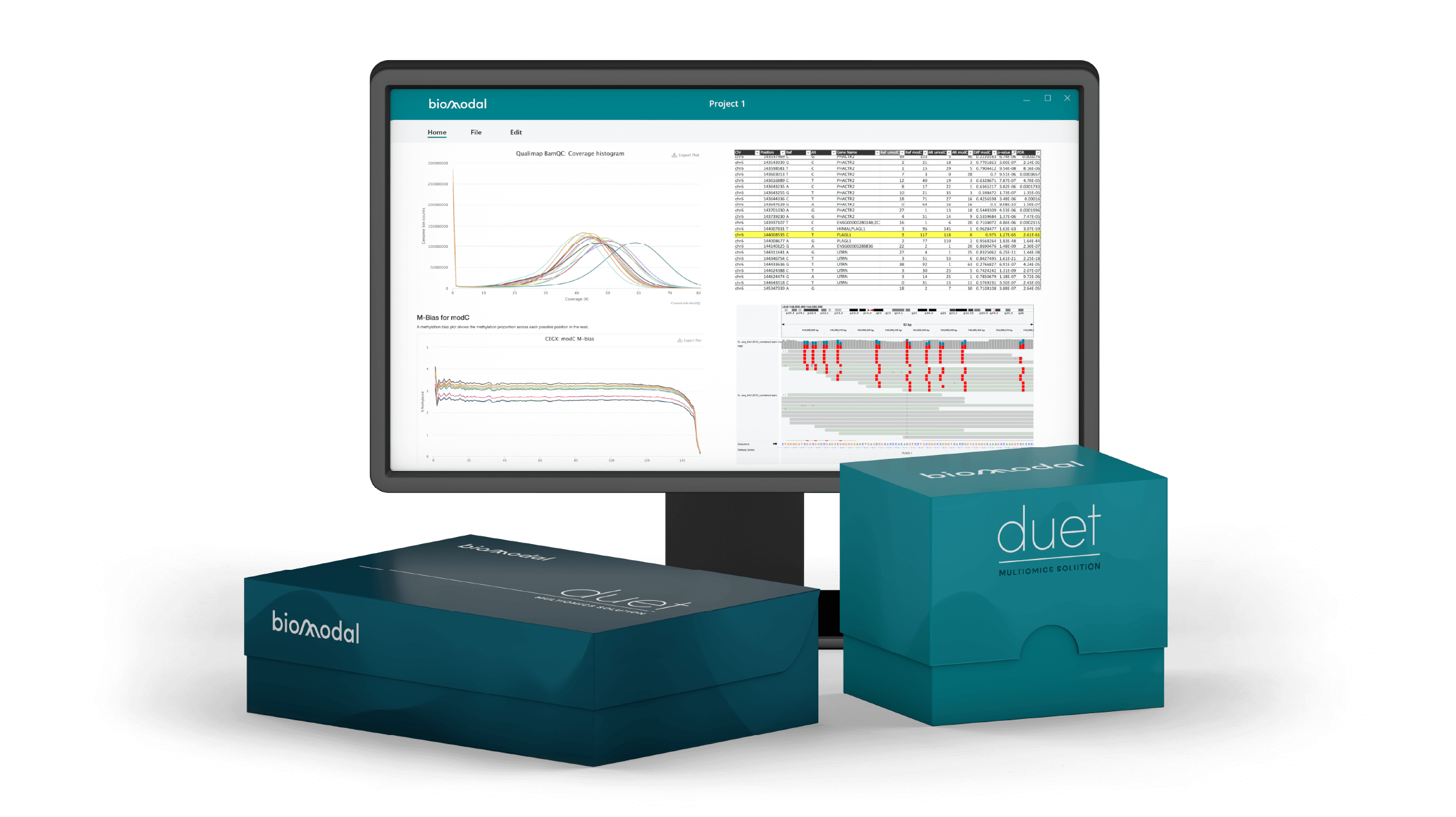
Early cancer detection has the potential to significantly improve treatment outcomes and survival rates. Epigenetic biomarkers in cell-free DNA, including DNA methylation, have been shown to differentiate between cancer and non-cancer and are already being integrated into liquid biopsy development programs. Traditional DNA methylation sequencing provides a conflated modified Cytosine (modC) readout, measuring CpGs that are 5-methylcytosine (5mC) or 5-hydroxymethylcytosine (5hmC) but not discriminating between the two states. Dynamic DNA demethylation occurs through TET enzyme activity, with conversion of 5mC to 5hmC preceding eventual loss of methylation. Hence, we hypothesized that obtaining separate measurements of 5mC and 5hmC would improve the ability to detect the development of colorectal cancer at the earliest stage.
We have therefore applied a technology which provides the 6-base genome (the complete genetic sequence whilst simultaneously distinguishing 5mC and 5hmC) from low nanogram input quantities of cfDNA. We generated whole genome 6-base data from cfDNA extracted from plasma of 32 healthy volunteers and 37 patients with colorectal cancer (CRC) at stages I and IV. We used machine learning approaches to build classifiers with features based on modC, 5mC alone, 5hmC alone, and both 5mC and 5hmC, to differentiate between cfDNA from patients with cancer and individuals without cancer, as well as between stage I CRC and Stage IV CRC.
Our findings indicate that separate measurements of 5mC and 5hmC significantly enhance diagnostic accuracy for the detection of stage I CRC (AUC = 0.95) compared to traditional approaches that conflate these markers (modified C, AUC = 0.66). Notably, most regions with an increase in 5hmC in stage I CRC cfDNA also decreased in 5mC in stage IV CRC, suggesting that 5hmC can effectively track regions undergoing demethylation during tumor development. These results support the hypothesis that distinguishing between 5mC and 5hmC can improve the sensitivity of liquid biopsy tests for early cancer detection.
We feel there is merit in applying 6-base sequencing to larger clinical cohorts, across different indications, to more broadly evaluate the potential of 6-base data to improve the earlier detection, diagnosis, and treatment of disease.