 |
 |
 |
 |
|---|
Documentation for duet software installation and running guide and the data interpretation guide has moved. Read the latest version of the documentation now.
Table of Contents
- 1.1. Download and unzip the complete set of installation and CLI scripts
- 1.2. Choosing the correct installation script
- 1.3. Setting up the pipeline on cloud VMs – Using the biomodal-cloud-utils script
- 1.4. Setting up the pipeline on HPC clusters
- 1.5. Import the duet multiomics solution pipeline using the biomodal CLI and perform a test run
- 1.6 Testing the duet pipeline on a larger dataset
- 1.7 nf-core community pipelines
1.1. Download and unzip the complete set of installation and CLI scripts
We have enabled a simple tool to directly download all the required script to your local computer, HPC node or cloud VM.
Please run the following command in a bash terminal window:
bash <(curl -s https://app.biomodal.com/cli/download)This script requires you to log in with your existing biomodal username and password.
If you do not have a biomodal account please contact us at support@biomodal.com
To reset the password for your account, please use this link https://app.biomodal.com/auth/pages/reset-password
The installation script require the following software modules:
| Dependency | Details |
|---|---|
| bash | version 4 or higher |
| jq (version > 1.6) | For JSON parsing |
| curl | Used for downloading software from the biomodal website |
| Google Cloud CLI | This should be a recent version that supports the gcloud storage command, including the required gcloud-crc32c libraries |
For the complete list of software dependencies, please refer to the section Installing pipeline and CLI software dependencies for details regarding dependencies on your platform.
On completing the curl command above, you will have downloaded the complete set of scripts as a zip file.
You have to unzip this file before you can start using the CLI scripts. Please note that you can use different tools to unzip the file, the example below use unzip but you can choose other tools, like jar xvf, to uncompress the file:
unzip biomodal.1.1.3.zipYou can now change directory to the unzipped biomodal folder and continue with the rest of the steps outlined in this guide.
1.1.1. Authentication tokens
Please note that the authentication process will add a new file to control the tokens required to authorise access to biomodal resources. The default location of this file is in the users $HOME directory and is called .biomodal-auth.json.
Each user running the CLI will have their own unique ${HOME}/.biomodal-auth.json file.
1.1.2. Shared log events
Downloading and running the biomodal CLI script requires you to log in at least once with your existing biomodal username and password. The CLI can optionally share runtime event notifications with the biomodal API service for the purpose of providing customer support.
The current event codes are being reported to the biomodal API services:
| Event code | Description | Optional? |
|---|---|---|
100 |
Downloaded the biomodal CLI package, via the biomodal API | No |
101 |
Completed running the biomodal init command |
Yes |
102 |
Completed running the biomodal test command |
Yes |
103 |
Completed running the biomodal analysis command |
Yes |
104 |
Completed running the biomodal dmr_call command |
Yes |
105 |
Completed running the biomodal reference make command |
Yes |
200 |
Customer prefer to automatically share events and metrics reports. Recorded once during the biomodal init step |
Yes |
201 |
Customer prefer to not share events and metrics reports automatically. Recorded once during the biomodal init step |
Yes |
202 |
User from the biomodal CRM system has registered for a software download and authentication account, via biomodal API | No |
Please note that the event codes are not shared with any third party and are only used for internal support purposes. The CLI version and the event code is shared via the biomodal API and will be registred against the user’s account in our internal CRM system.
If you do not wish to share the optional events, please decline event and metrics report sharting during the biomodal init stage, or manually set share_metrics: false in the biomodal CLI configuration file, config.yaml. Please see section “1.4.2.2. CLI config file” below for more information.
If you opt out of sharing events and metrics reports, the CLI will not share any events or metrics reports. However, you will still have to authenticate with the biomodal API service to download biomodal software, test data and reference data.
Nextflow will automatically check for new software versions. To disable this feature please additionally set the NXF_DISABLE_CHECK_LATEST enviroment variable to true in the shell environment where you plan to run the duet pipekine. This will disable the automatic version check for the Nextflow software.
export NXF_DISABLE_CHECK_LATEST=trueIf you do not have a biomodal account or like to reset your password, please contact us at support@biomodal.com
1.1.3. Required permissions for cloud tenancies
We recommend that a least privilege approach is taken when providing users with permissions to create cloud resources. The cloud specific examples below demonstrate minimum required permissions to bootstrap resources for aws and
gcp environments.
AWS
We recommend an administrator runs ./bimodal-cloud-utils create aws to minimise potential permissions issues. Please review this document for specific policies and roles required to run the biomodal-cloud-utils utility for AWS.
bimodal publish a specific AWS Terraform module in a GitHub repository located here: https://github.com/cegx-ds/terraform-aws-bootstrap.git. Organisations may not allow public GitHub repositories to be accessed without “whitelisting” these first. If you encounter an error similar to the one below, please contact your IT department to allow access to this repository by running the command referenced below. Make sure you replace the /biomodal/ reference with the current installation location you are using for the biomodal CLI.
Could not download module "bootstrap" source code from "git::https://github.com/cegx-ds/terraform-aws-bootstrap.git ": error
downloading 'https://github.com/cegx-ds/terraform-aws-bootstrap.git': /usr/bin/git exited with 128: fatal: detected dubious ownership in repository at '/biomodal/terraform/aws/.terraform/modules/bootstrap'
To add an exception for this directory, call:
git config --global --add safe.directory /biomodal/terraform/aws/.terraform/modules/bootstrapNetwork considerations for AWS
The newly bootstrapped environment will assume AWS Batch instances are able to automatically assign public IP addresses to allow internet egress.
This feature can be manually enabled in the console, via the Auto-assign public IPv4 address option in the subnet’s configuration. If disabled, instances deployed by AWS Batch will fail to be able to pull required external images and AWS Batch jobs will ultimately fail.
If this option must be disabled, you should ensure a NAT gateway is deployed within the selected VPC that allows instances within the subnet to access images from ECR and quay.io.
GCP
We recommend using GCP’s predefined roles. These roles are created and maintained by Google so you do not need to create custom roles.
Below are the recommended predefined roles you require access to create cloud resources for running this CLI.
| Role Name | Purpose | Required |
|---|---|---|
roles/storage.admin |
If a new bucket is required | No (unless new bucket is specified) |
roles/artifactregistry.writer |
Create required artifact registry | Yes |
roles/iam.serviceAccountCreator |
Creating required service account(s) | Yes |
roles/compute.admin |
Creating required compute resources and instance template | Yes |
roles/iap.admin |
Allowing IAP access to VMs | Yes |
roles/resourcemanager.projectIamAdmin |
To assign project wide roles to service accounts | Yes |
roles/storage.legacyBucketWriter |
Allow read/write permission on existing bucket | No (Required if wishing to use an existing bucket) |
Conditions and custom roles can also be created for more granular access restriction, however this is beyond the scope of this document.
1.1.4. Software dependencies
Please refer to the section Installing pipeline and CLI software dependencies for details regarding software dependencies on your platform.
(back to main documentation) | (back to top)
1.2. Choosing the correct installation script
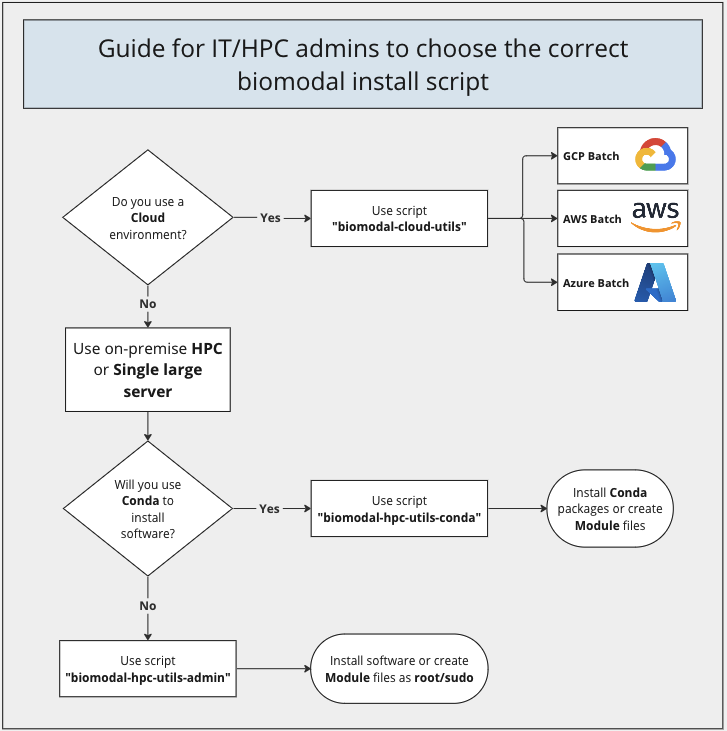
(back to main documentation) | (back to top)
1.3. Setting up the pipeline on cloud VMs – Using the biomodal-cloud-utils script
We have created a bootstrap tool to assist with the creation of your cloud resources required to run the duet pipeline with Nextflow. The 3 currently supported clouds are gcp, aws and azure.
The biomodal-cloud-utils utility is intended for DevOps / System administrators that create and maintain Cloud tenancies. It utilises Terraform as the infrastructure and code software tool that will create all the compute resources needed to run biomodal pipelines with Nextflow. Please see the respective documentation for your cloud provider for more info, referenced later in this guide.
Please follow the following steps from a Linux compatible computer:
-
Install Terraform – see this link for OS-specific instructions.
-
Make sure your cloud admin has given your account enough privileges to create cloud resources over API (see required permissions).
-
Authenticate Terraform by your cloud provider:
3.1. Authenticate Terraform – GCP
3.2. Authenticate Terraform – AWS
3.3. Install Azure CLI and sign in to your Azure account
-
Execute the following command to create a new cloud environment from the provider of choice:
./biomodal-cloud-utils create <cloud> # gcp / aws / azureNote: If the resource creation process is interrupted, please use the
destroycommand before attempting to usecreateagain. -
Execute the following command to connect to the resources created in the above steps:
./biomodal-cloud-utils connect <cloud> # gcp / aws / azureNote: Please wait 4-5 more minutes after the
createcommand has completed to allow the cloud resources to fully initialise.You can now continue with section 1.5. Import the duet multiomics solution pipeline using the biomodal CLI and perform a test run.
-
To destroy the resources you created, execute:
./biomodal-cloud-utils destroy <cloud> # gcp / aws / azureWARNING: Destroying your resources is a permanent action, only run this command when you are sure you no longer need them. Non-empty buckets should not be deleted.
The following command is designed for testing your Terraform config and can be use for CI-CD actions:
./biomodal-cloud-utils test <cloud> # gcp / awsYou can also pass in the --remote argument to automatically generate a remote backend.
./biomodal-cloud-utils test <cloud> --remoteYou can provide variables to the test command via Terraform environment variables (i.e. TF_VAR_foo). You can also pass in backend remote config via environment variables.
Please note that the cloud VM will be created with a public IP to allow for data ingress/egress. Please update this as required after software install, authentication, configuration and testing.
(back to main documentation) | (back to top)
1.4. Setting up the pipeline on HPC clusters
Please do not attempt installing the CLI or duet pipeline without extensive knowledge of the local Linux HPC environment. We recommend that you contact your local HPC admin or IT support before attempting to install the CLI or duet pipeline. Restrictive quotas on local disk resources and software install limitations are likely to cause issues during installation and update operations.
The software installation scripts described in this section are not intended to be re-run when you upgrade to a newer version of the biomodal CLI or duet pipeline. Please see section 5.2 Manual update biomodal CLI script from 1.0.x to 1.1.3 for more information on how to update the CLI and pipeline software.
1.4.1. Using the biomodal bootstrap on HPC tool
We have created a bootstrap tool to assist with installing required software and downloading the pipeline on an HPC cluster, or standalone server. Currently, Ubuntu and CentOS/RHEL systems are supported. Other Linux distributions may work fine providing they use apt-get or dnf package managers, but we have not tested them.
The biomodal-hpc-utils-admin script will install the required OS packages and software locally to run a Nextflow pipeline or workflow with sudo access. The suggested HPC setup will use Singularity as an alternative to Docker. Please note that Singularity does not enable nor propagate root privileges to the Singularity images.
NB!! Before you execute the biomodal-hpc-utils-admin script, make sure your account has sudo privileges. The biomodal-hpc-utils-admin script will require sudo access and must be reviewed by an HPC admin before executing as every HPC installation will require different settings. This script will create all the local folders and settings files for the pipeline, install required OS & system software, and download the duet pipeline software.
There is an alternative script to install software without using root or sudo access. This script is called biomodal-hpc-utils-conda. This should not be used by a non technical user as extensive knowledge of the local Linux HPC environment is required.
Please review each step in the biomodal-hpc-utils-admin and biomodal-hpc-utils-conda script(s) before you run it on any HPC cluster or local machine. Do not assume that all steps in these scripts will be an exact match for your local server or HPC nodes. If you choose to use the biomodal-hpc-utils-conda script, we recommend you use an existing working conda environment and have the required permissions to install software in that environment.
Executing the biomodal-hpc-utils-admin or biomodal-hpc-utils-conda scripts will require in-dept knowledge of the local Linux HPC cluster environment and should not be attempted by a non technical user.
Executing the scripts will ask to confirm that you have read the documentation and understand the implications of running this script on your system.
Please confirm that you have read the documentation and understand the implications of running this script on your system.
Are you sure you want to run this script? (y/n)The rest of section 1.4 in this guide will only refer to the biomodal-hpc-utils-admin script. We recommend using a terminal multiplexer to manage your persistent shell sessions, such as tmux to ensure long running sessions are not terminated.
To initiate installation, execute the following command:
./biomodal-hpc-utils-adminThe script will configure software and create two local files that the main biomodal tools will use to local data directories and other resources. Please follow the following prompts and select accordingly:
Enter cli installation path. : /biomodalYou can leave this as default or select an installation path of your choosing.
Enter data bucket path - shared location where ref data and images will be stored. : /biomodal/data_bucketYou can leave this as default or select a data storage path as default storage directory for reference data, input, and output.
Choose relevant geographical location of the biomodal docker registry (1-3)?
1) asia
2) europe
3) us
#?You need to select a region, from which biomodal GCP resources will be pulled from.
Current queueSize is: 200
This is the maximum number of concurrent jobs that the pipeline will attempt to launch
Would you like to change it? (y/n)You can limit the maximum number of concurrent Nextflow jobs that the duet pipeline will attempt to launch. This is useful for HPC cluster or Cloud projects with limited resources.
Which executor would you like to use? (choose 1-4)
1) slurm
2) lsf
3) sge
4) local
#?Please select the Nextflow executor of choice; if you are not using a HPC cluster scheduler, please select local.
The local executor setting will still require a server with significant memory, CPU and disk space to run the pipeline.
We recommend using terminal multiplexer to manage your persistent shell sessions, such as tmux. This is due to the potentially long running nature of pipelines.
When the script runs to completion, please log out and back in again to activate any new environment variables. There may be a requirement on your local node to restart services due to OS package updates.
If you want to edit the config and/or Nextflow files manually, please see details below.
(back to main documentation) | (back to top)
1.4.2. Installing duet pipeline and software manually
1.4.2.1. Installing pipeline and CLI software dependencies
If you are using an unsupported OS, do not have sudo access or are unable to use the biomodal-hpc-utils-admin tool, please install and/or check if the following dependencies are installed in your environment:
| Dependency | Details |
|---|---|
bash |
version 4 or higher |
| JAVA | version 8 or higher |
| Nextflow | A recent version is recommended, tested and supported from 21.06 onwards |
| Google Cloud CLI | Please ensure you have a recent version as this is used for software installation and authentication. Only required on the nodes that install the pipeline and workflow software, is not part of running the duet pipeline. |
| Singularity | Aka Apptainer. Please note that Apptainer is only supported by more recent versions of Nextflow, so we recommend checking if your version supports Singularity and/or Apptainer |
tar, unzip |
Used to uncompress the duet pipeline, reference data and CLI software from biomodal |
rsync |
Copies files internally on HPC platforms |
pcre2grep |
Used to search for patterns in text files |
curl |
Download software from biomodal.com |
jq |
For JSON parsing |
| Other OS support packages: | gnupg, lsb-release, apt-transport-https, gnupg, ca-certificates |
Some installations may additionally require Google SDK components to ensure integrity of data being downloaded, this can be installed using: gcloud components install gcloud-crc32c
If you are missing any of the above, please contact your HPC or IT administration for installation/enablement of these packages.
You can refer to the biomodal-hpc-utils-admin script as guide to install these packages on different platforms.
1.4.2.2. CLI config file
The biomodal CLI script relies on a config file called config.yaml. The biomodal-hpc-utils-admin script will add this file to the default location, /biomodal/, or another user provided location. This config file is keeping track of the current version of the pipeline (duet_version) and the pipeline data location (directory or bucket_url).
If you are not using the biomodal-hpc-utils-admin, please create the file with the following content.
platform: hpc
duet_version: 1.4.1
ref_pipeline_version: 1.0.1
bucket_url: <default bucket/folder location of your pipeline data input / output>
work_dir: <folder location/path where pipeline temporary data is stored>
init_folder: <folder location/path where duet pipeline software is stored>
biomodal_registry: <biomodal_registry, see below>
biomodal_api_baseurl: https://app.biomodal.com
biomodal_releases_bucket: gs://cegx-releases
biomodal_reference_bucket: gs://release-reference-files
share_metrics: false
error_strategy: normalinit_folder should point to the location of the duet pipeline and D(h)MR workflow software folder and work_dir should point to the temporary/scratch location for the duet pipeline. if work_dir is not added, the default <bucket_url>/nf-work/ location will be used when running the duet pipeline.
Please note that the location you choose for work_dir should have about 6 to 7 times the size of the data in your --input-path location in free space to avoid running out of disk space. Additionally, if the work_dir is located on the same disk as the location you choose for the --output-path and --input-path parameters, you should aim to have about 10 times the input space in free space before you start the analysis.
The biomodal_registry should be one of the following:
asia-docker.pkg.dev/cegx-releases/asia-prodeurope-docker.pkg.dev/cegx-releases/eu-produs-docker.pkg.dev/cegx-releases/us-prod
share_metrics should be true or false. If set to true, events and the duet pipeline metrics report will be shared with biomodal after a successful analysis run. If set to false, events and the duet pipeline metrics report will not be shared with biomodal after a successful analysis run.
Please make sure that the biomodal script is in the same location as the config files.
1.4.2.3. Nextflow config file
Nextflow relies on the nextflow.config file in the biomodal script folder so that users can adjust the process section to match any local HPC scheduler and queue setup.
This local configuration file is applied after any settings defined in the standard duet pipeline configuration files. Nextflow will automatically cascade the settings in the duet pipeline configurations, followed by the nextflow.config in the biomodal script folder. This ensures that your customised settings are always retained in the nextflow.config file in the biomodal script folder when upgrading to any new releases of the duet pipeline.
If you are not using the biomodal-hpc-utils-admin script, please create the nextflow.config file with the following content.
singularity {
enabled = true
autoMounts = true
libraryDir = “<the location you used in the config.yaml file>"
}
params {
registry = "<biomodal registry from choices below>"
}
process {
executor = "slurm"
}
report {
overwrite = true
}Note: Please fill the “registry” value under “params” with one of the following:
"asia-docker.pkg.dev/cegx-releases/asia-prod""europe-docker.pkg.dev/cegx-releases/eu-prod""us-docker.pkg.dev/cegx-releases/us-prod"Note: Please fill the “process” section according to your queue setup (select local if no HPC scheduler is present) to one of the following:
process {
executor = "slurm"
}process {
executor = "lsf"
}process {
executor = "sge"
process.penv = "smp"
clusterOptions = "-S /bin/bash"
//If your system is not supprting h_rt/h_rss/mem_free settings, you can try to use the following settings
//clusterOptions = { "-l h_vmem=${task.memory.toString().replaceAll(/[\sB]/,'')}" }
}process {
executor = "local"
}These settings should be adjusted to match your HPC setup. For more advanced settings, please see 7. HPC Recommendations
(back to main documentation) | (back to top)
1.5. Import the duet multiomics solution pipeline using the biomodal CLI and perform a test run
After you have created a new cloud or HPC environment, you can start using the biomodal CLI tool.
To see all the command and options you can use with the biomodal CLI tool, simply run biomodal with no parameters.
The biomodal CLI will:
- Import the latest version of the duet pipeline and D(h)MR workflow.
- Import required reference files, for both the duet pipeline and Reference Generation pipeline.
- Import the biomodal test data set, aka. ‘RunXYZ’, to make sure all the resources created manually or with Terraform work as expected.
- Synchronise all required biomodal Docker images from our host repository to your project repository.
- Enable you to update to a new version of the duet pipeline and reference data when available.
- Do a test run with biomodal test data set to make sure everything works as expected with Nextflow and configuration.
Note: When using a cloud environment, please make sure that you can launch Terraform and be authenticated with your cloud provider either from your local machine, a Cloud Shell, or existing VM in your cloud provider (see section 1.2.).
Note: The CLI scripts and required configuration files are available in the /biomodal folder. This is also added to your $PATH variable (when biomodal-cloud-utils or biomodal-hpc-utils-admin are executed). If you used an alternative location on HPC, please refer to the biomodal using the full path to its location.
On your VM or HCP command line, run biomodal init
This will transfer the pipeline, reference files, test data set, and Docker images (Singularity on HPC) to the data location specified in the config.yaml file. If the data download process is interrupted, please remove data under the reference_files folder and re-run biomodal init.
The duet pipeline will store temp files in a separate cache directory. This is a folder that will typically contain a larger amount of files and can be cleaned out after successful analysis runs.
The duet pipeline temp directory is currently set as <current temp folder>.
Press Enter to continue using this directory or enter a new one here: <new temp folder>You will be asked if you want to automatically upload the pipeline metrics report to biomodal after a successful analysis run. This will enable biomodal to compare your pipeline setup with the biomodal internal pipeline for the purpose of quality control. This will share the metrics CSV report and the parameters you used to run the pipeline, no other data will be shared.
If you do not want to share the report automatically, you can manually run biomodal report <metrics report file name> <pipeline json parameters file name> to share a specific metrics report at any later time.
Would you like to share the pipeline metrics report at the end of all successful analysis runs with biomodal? (1-2)
1) Yes
2) No
#?You can specify which location you prefer the duet pipeline software to be installed to. This may be in your $HOME folder or on a shared software filesystem. Please note the directory must be created before running biomodal init.
The duet software location 'init_folder' property is not currently defined in the configuration file
Would you like to set it to /<home_folder>? (1-2)
1) Yes
2) No
#?You will now be asked about the duet pipeline error strategy you would like to use:
- Normal: Retries failed jobs up to 10 times depending on the exit status. This option will retry all failed jobs and continue with other samples should there be a problem with a specific sample.
- FailFast. Retries failed jobs only once or exits immediately depending on the exit status and the executor.
Please select which duet pipeline error strategy you would like to use:
1) Normal.
2) FailFast.
#?When biomodal init has completed, you can run biomodal test to test the pipeline using biomodal test data (Aka. “RunXYZ”).
The test run is expected to complete similar to the below example.
Completed at: 10-Mar-2023 12:51:33
Duration : 45m 15s
CPU hours : 10.0
Succeeded : 27(back to main documentation) | (back to top)
1.6 Testing the duet pipeline on a larger dataset
The biomodal test command will verify that all the software necessary to run the duet pipeline works as expected on a small test sample. However, it can be useful to run another test on a larger dataset, which is more representative of the scale of data generated by a biomodal assay. To perform this larger test, we will use the publicly available Genome In A Bottle (GIAB) data published by biomodal.
Firstly, go to the demo dataset instructions, and follow the steps in the section entitled “Download Instructions” to download fastq files just as they would look when generated by a sequencing instrument. You can either download the +modC or evoC sample data – either will provide a good test of the duet pipeline – just make sure you select the appropriate option when running the biomodal analyse command.
Then, assuming you have downloaded the raw fastqs to a directory called $BIOMODAL_PATH/giab_data/nf-input, run the following command:
biomodal analyse \
--input-path $BIOMODAL_PATH/giab_data \
--meta-file CEGX_Run_meta.csv \
--output-path $BIOMODAL_PATH/output \
--additional-profile deep_seq \
--tag giab_demo_data \
--mode 5bpNote that in this case, the --mode 5bp parameter has been used – this needs to match the type of data you downloaded.
(back to main documentation) | (back to top)
1.7 nf-core community pipelines
The nf-core community has a number of publicly available Nextflow pipelines. The nf-core pipelines are designed to be easy to use and are well documented. You can find a list of the nf-core pipelines at https://nf-co.re/pipelines. Some nf-core pipelines may have a similar technical setup as your pipeline execution enviroment, so it is worth checking if there are relevant Nextflow pipeline settings and parameters you can apply to your nextflow environment.
