biomodal
- Casper K Lumby
- Nicholas Harding
- Jamie Scotcher
- Shirong Yu
- Páidí Creed
- Joanna D Holbrook
Broad Institute
- Casper K Lumby
- James Emery
- Michael Gatzen
- Christopher Kachulis
- Megan Shand
- Eric Banks
There is more to DNA than the genetic alphabet A, C, G and T. Epigenetics plays a causal role in cell fate, ageing and disease development. Methylated cytosines, such as 5mC and 5hmC, represent important biomarkers and are informally considered the 5th and 6th bases of DNA:
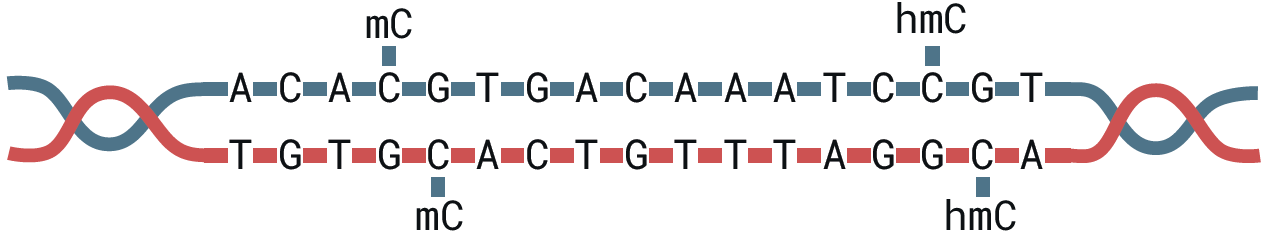
The combination of genetics and methylation has proved to be more powerful than either modality on their own. However, current methylation detection technologies rely on sacrificing genetic information for epigenetic in-sight:
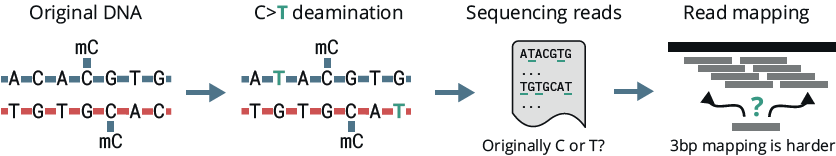
We present a novel sequencing technology, duet multiomics +modC, that jointly deter-mines genetics and methylation at high accuracy. In this poster we examine the genetic accuracy of the technology and benchmark it against existing methylation detection methods. This work derives from a collaboration between biomodal and the Broad Institute.
The Genome in a Bottle (GiaB) Consortium provides a complete genetic charac-terisation of 7 human samples (HG001-HG007). We sequenced all seven sam-ples in two replicates across four technologies: Whole-genome sequencing (WGS), whole-genome bisulfite sequencing (WGBS), Enzymatic Methyl-seq (EM-Seq) and 5-Letter seq.
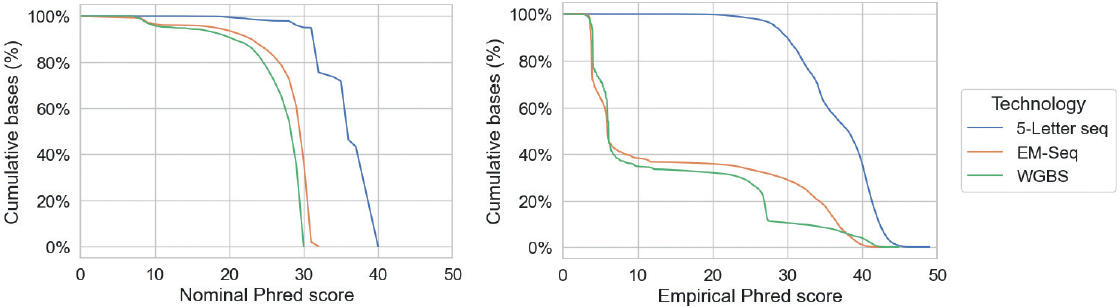
Phred scores describe the accuracy of a base call, e.g. Q30 means that a base is 99.9% certain to be correctly called. We make two distinctions:
- Nominal Phred scores: These are accuracy estimates provided by the sequencing instrument. These may not be 100% accurate.
- Empirical Phred scores: These are accuracy evaluations obtained by comparing called bases with known bases.
Below are nominal and empirical Phred distributions. About 90% of 5-Letter seq bases have a Phred score greater than Q30 and around 35% have a score larger than Q40:
We can further stratify genetic accuracy by base type and GiaB sample:
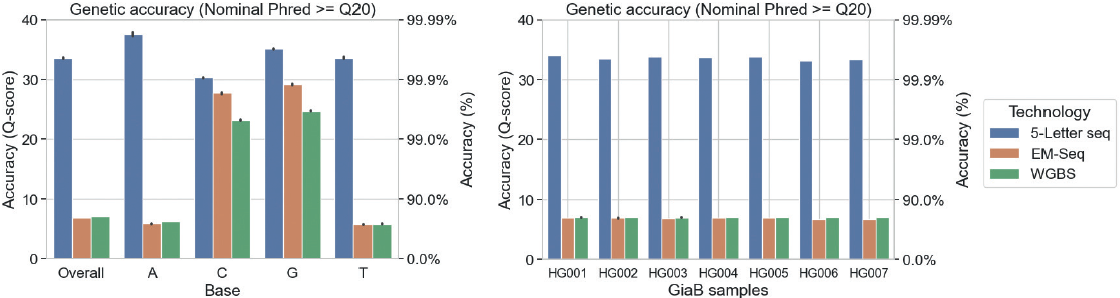
The accuracy of EM-Seq and WGBS is lower than that of 5-Letter seq. This is driven by C>T deamination, which results in reduced accuracy for T (forward strand) and A (reverse strand) bases, and read mapping using only 3 bases. Genetic accuracy is consistent across all 7 GiaB samples.
SNP calling was performed using GATK4 for 5-Letter seq and using Bis-SNP for EM-Seq and WGBS. Evaluation showed that 5-Letter seq was significantly more accurate at variant calling than EM-Seq and WGBS:
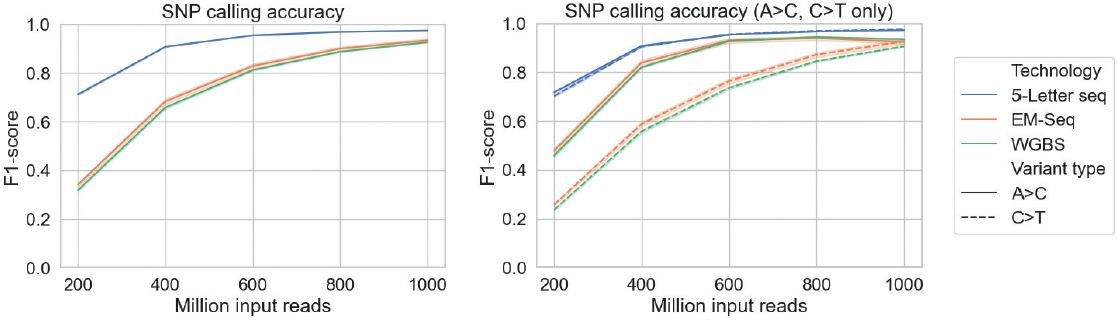
Additionally, 5-Letter seq performance was independent of SNP variant type:
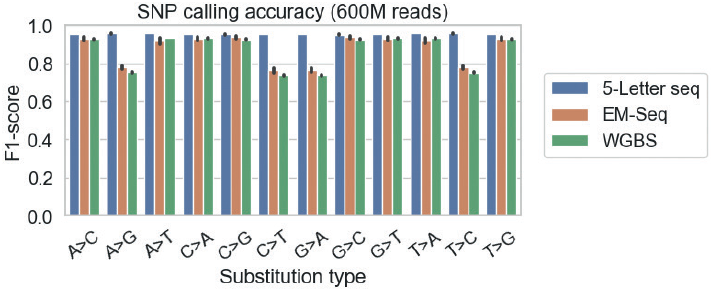
| Impacted by C>T deamination | Variant type |
|---|---|
| Yes | C>T, T>C, A>G, G>A |
| No | A>C, A>T, C>A, C>G, G>C, G>T, T>A, T>G |
With currently available methods, to achieve simultaneously high genetic and epigenetic accuracy, it is necessary to perform two separate workflows. This approach is limited by sample availability and the need for phasing data:
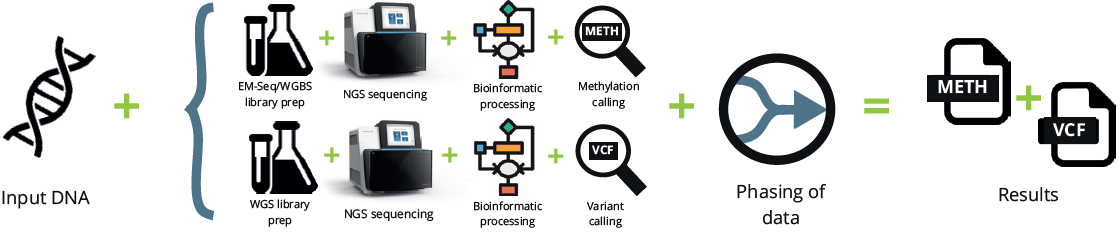
Under this setup, WGS variant calling is based on half the total sequencing volume. With 5-Letter seq, the above can be achieved with a single workflow. However, 5-Letter seq accomplishes this by resolving two reads into one. Therefore, SNP calling is compared here on coverage rather than number of input reads:
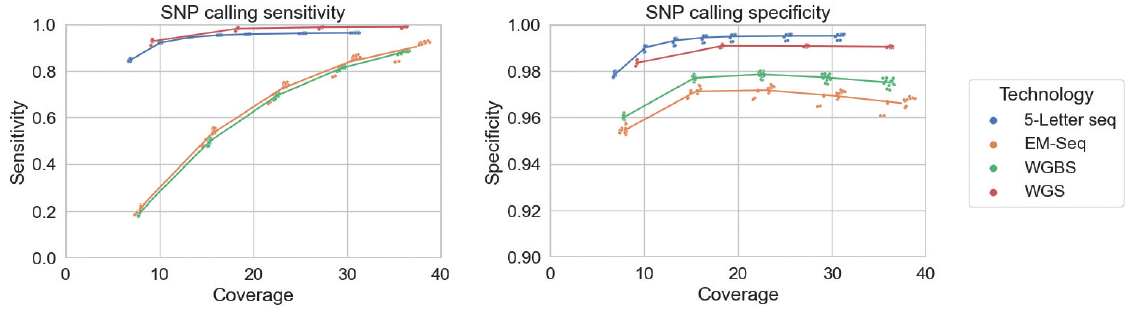
5L seq is more specific (0.5% more at 20X) and less sensitive (2.6% less at 20X) than WGS. Overall, 5-Letter seq produces highly accurate genetic and epigenetic calls. The phased nature of the data allows for generating novel insights (see other posters).