- Fabio Puddu¹
- Annelie Johansson¹
- Aurélie Modat¹
- Jamie Scotcher¹
- Riccha Sethi¹
- Shirong Yu¹ ²
- Nick Harding¹
- Mark S. Hill¹
- Ermira Lleshi¹
- Casper Lumby¹
- Jean Teyssandier¹
- Michael Wilson¹ ³
- Robert Crawford¹
- Tom Charlesworth¹
- Robert J Osborne¹
- Shankar Balasubramanian¹ ⁴ ⁵
- Páidí Creed¹
1 biomodal Ltd, The Trinity Building, Chesterford Research Park, Cambridge, UK.
2 Current address: Tagomics Ltd, The Cori Building, Little Abington, Cambridge, UK.
3 Current address: Department of Astrophysical Sciences, Princeton University, New Jersey, US.
4 Cancer Research UK Cambridge Institute, University of Cambridge, Cambridge, UK.
5 Yusuf Hamied Department of Chemistry, University of Cambridge, Cambridge, UK.
Early cancer detection has the potential to significantly improve treatment outcomes and survival rates. This study investigates the roles of 5‑methylcytosine (5mC) and 5‑hydroxymethylcytosine (5hmC) as biomarkers for early-stage colorectal cancer (CRC) detection in cell-free DNA (cfDNA). Using duet evoC for whole genome 6-base sequencing¹, we analyzed cfDNA from 37 treatment-naive CRC patients and 32 healthy controls.
Our findings indicate that combining measurements of 5mC and 5hmC significantly enhances diagnostic accuracy (AUC = 0.95) compared to traditional approaches that conflate these markers into a modified cystosine call (modC, AUC = 0.66).
Notably, 71.7% of differentially methylated regions (DMRs) exhibiting an increase in 5hmC in stage I cfDNA also showed a corresponding decrease in 5mC in stage IV, suggesting that 5hmC can effectively track regions undergoing demethylation during tumor development.
These results support the hypothesis that distinguishing between 5mC and 5hmC can improve the sensitivity of liquid biopsy tests for early cancer detection.
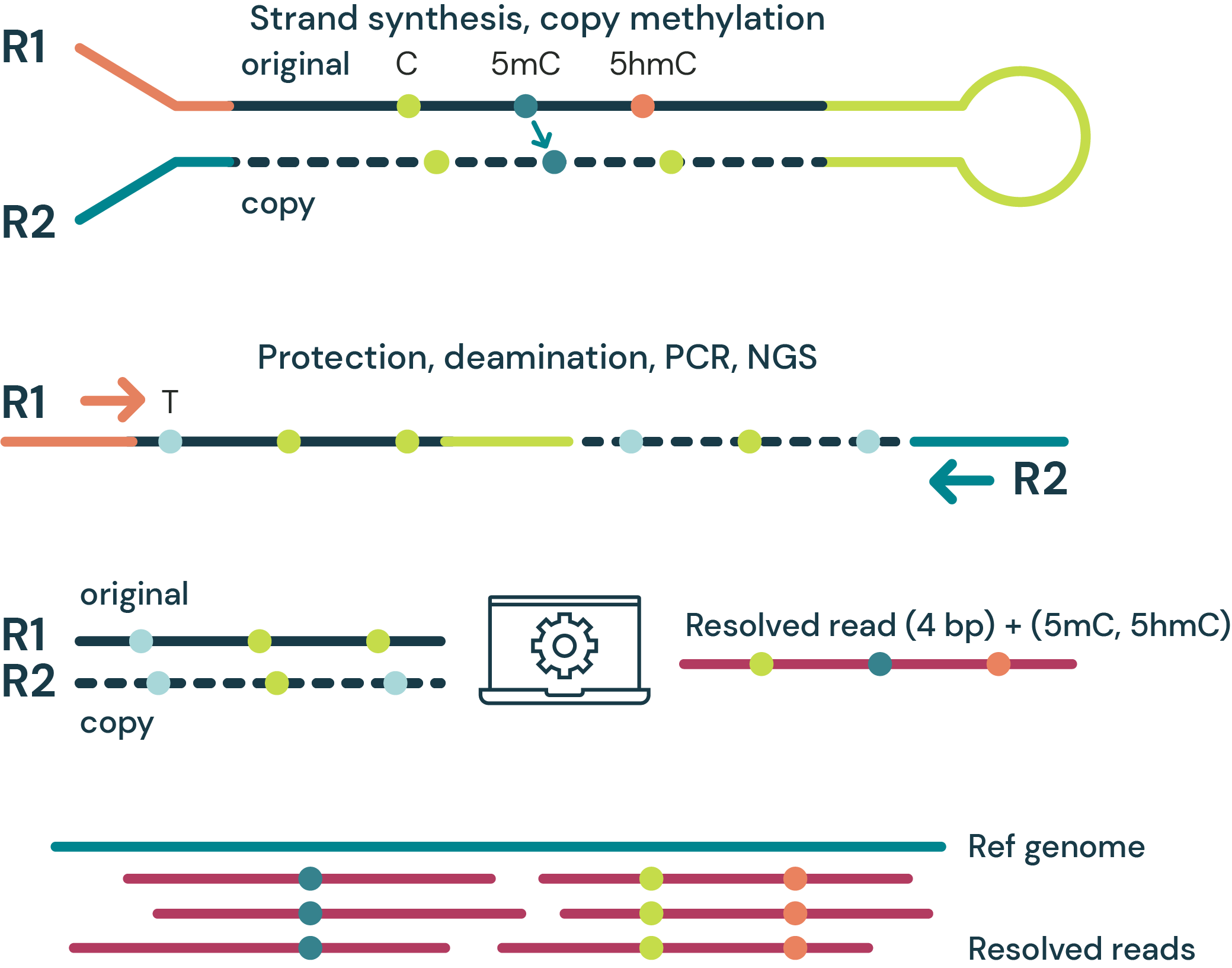
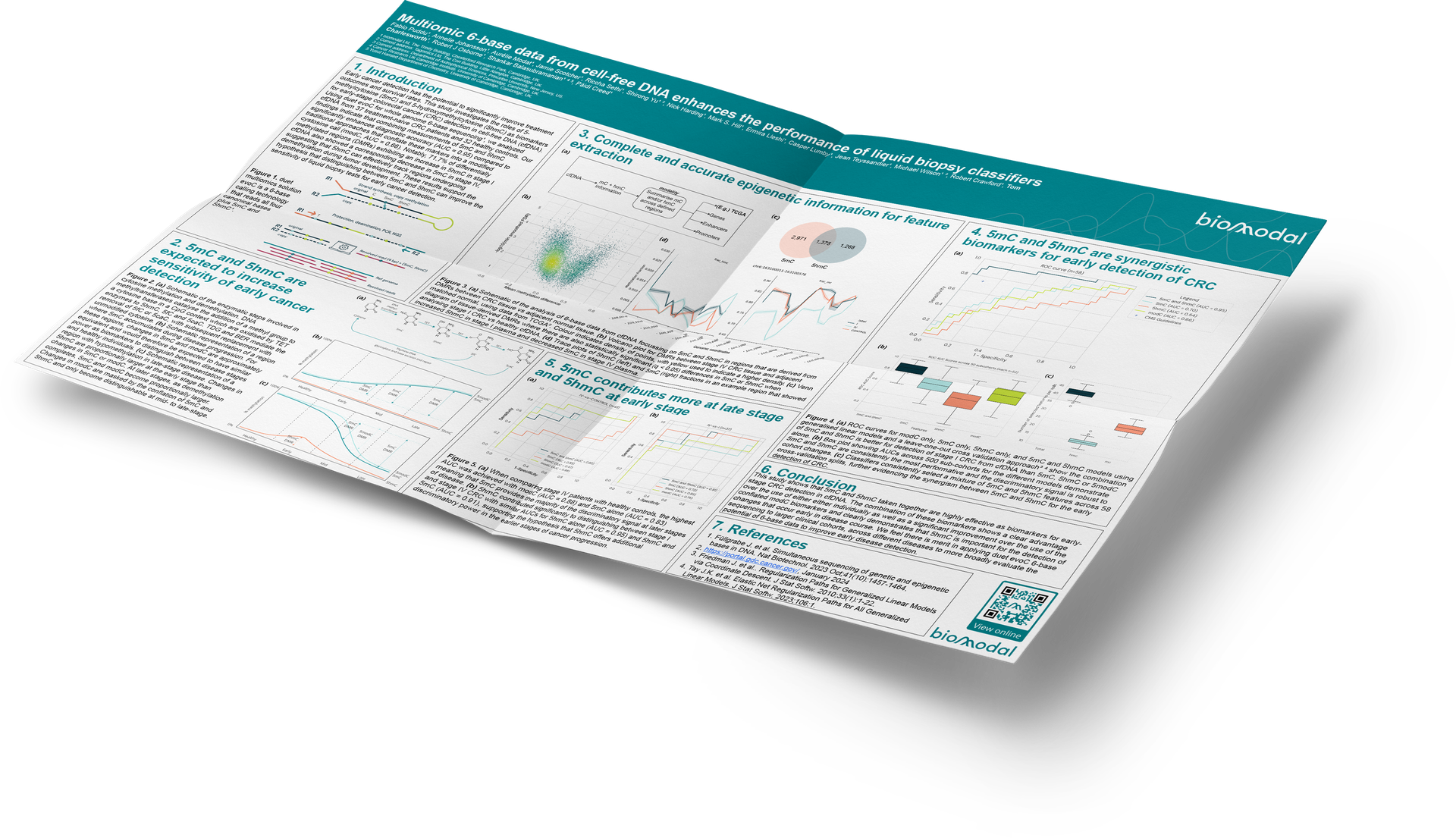
Figure 1. duet multiomics solution evoC is a 6-base calling technology that reads all four canonical bases plus 5mC and 5hmC¹.
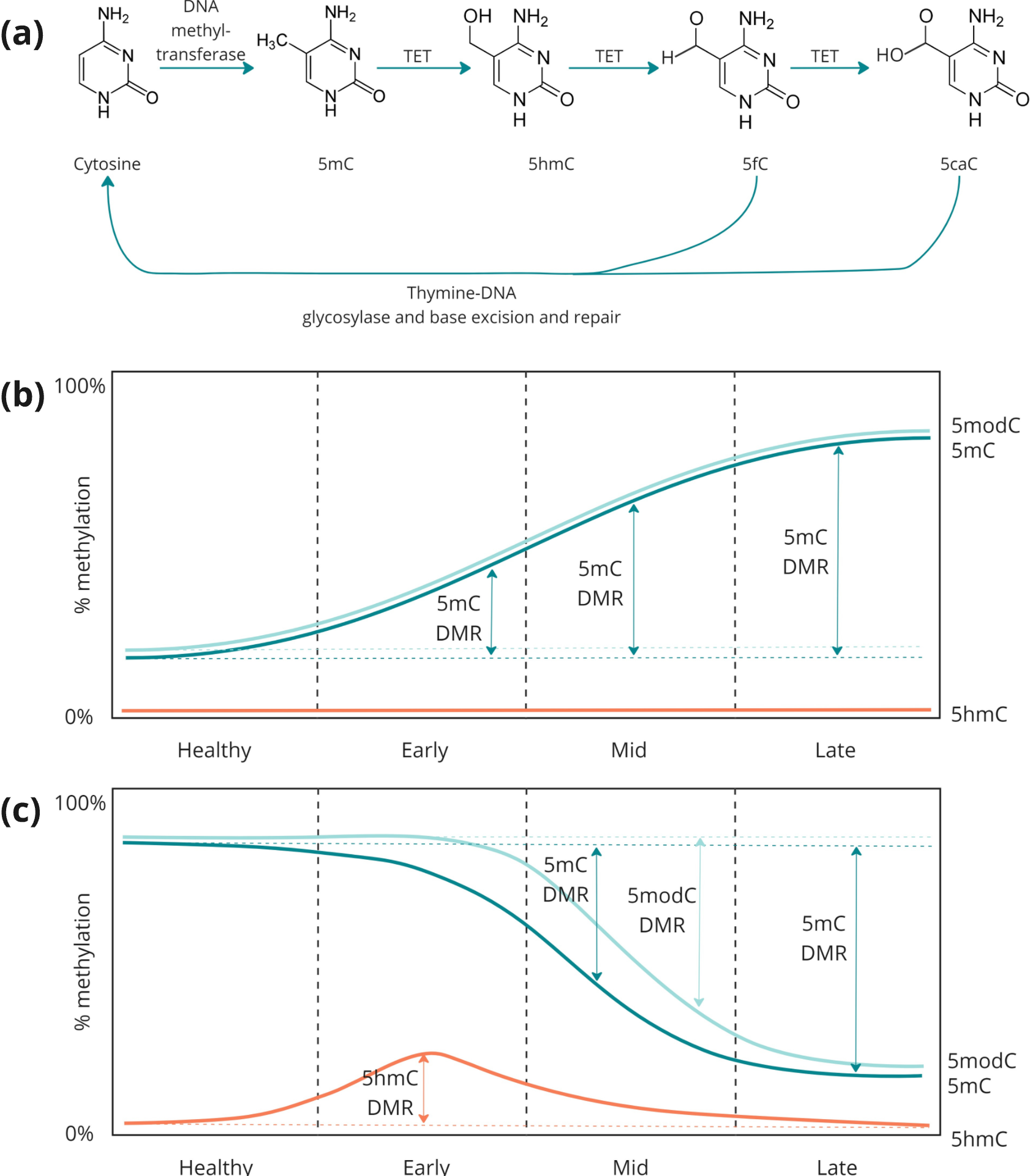
Figure 2.
- Schematic of the enzymatic steps involved in cytosine methylation and demethylation. DNA methyltransferases catalyse the addition of a methyl group to a cytosine base in a CpG context which are oxidised by TET enzymes to 5hmC, 5fC and 5caC. TDG and BER mediate the removal of 5fC or 5caC, with subsequent replacement with unmodified cytosine.
- Schematic representation of a region where 5mC accumulates during disease progression. For these regions, changes in 5mC and modC are approximately equivalent and would therefore be expected to have similar power as biomarkers to distinguish between disease stages and healthy individuals.
- Schematic representation of a region with hypomethylation in late-stage disease. Changes in 5hmC are proportionally larger at the early stage than changes in 5mC or modC. At later stages, as demethylation completes, 5mC and modC become proportionally larger. Changes in modC are masked by the conflation of 5mC and 5hmC and only become distinguishable at mid- to late-stage.
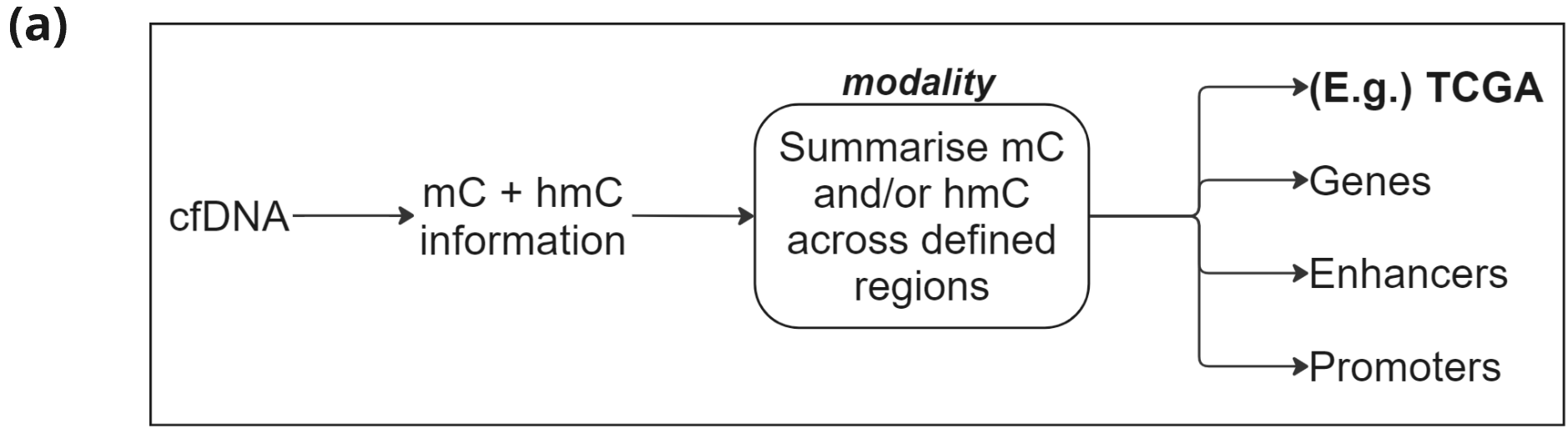
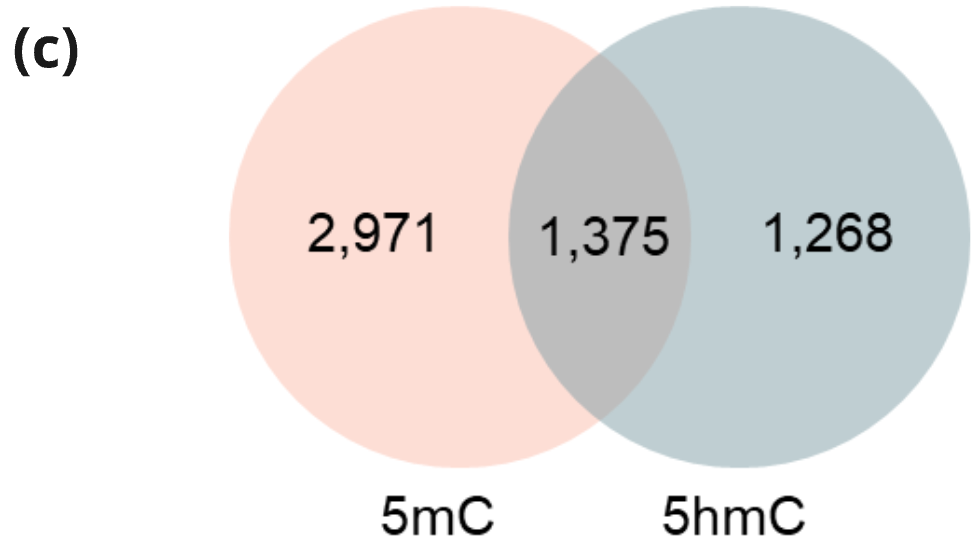
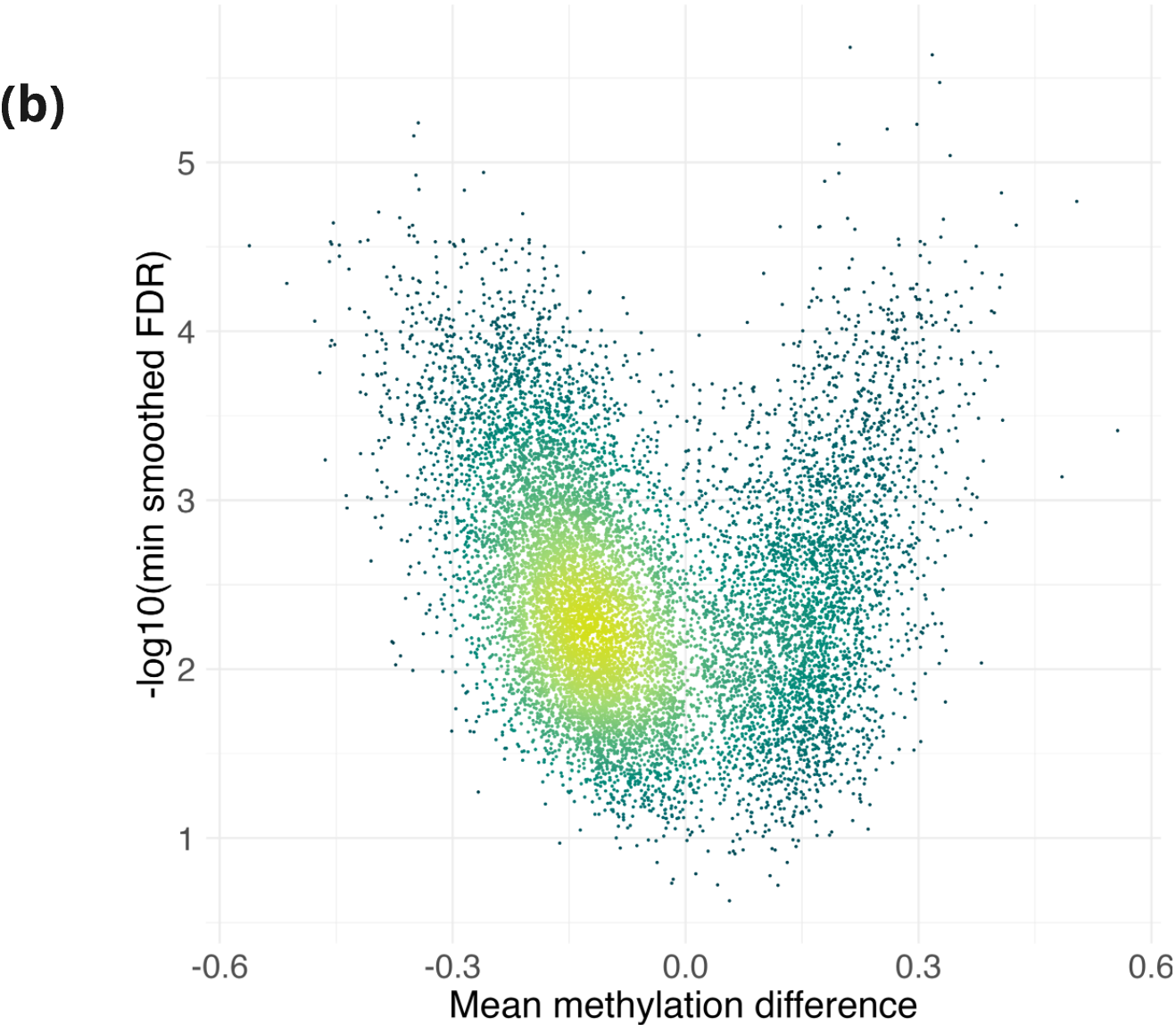
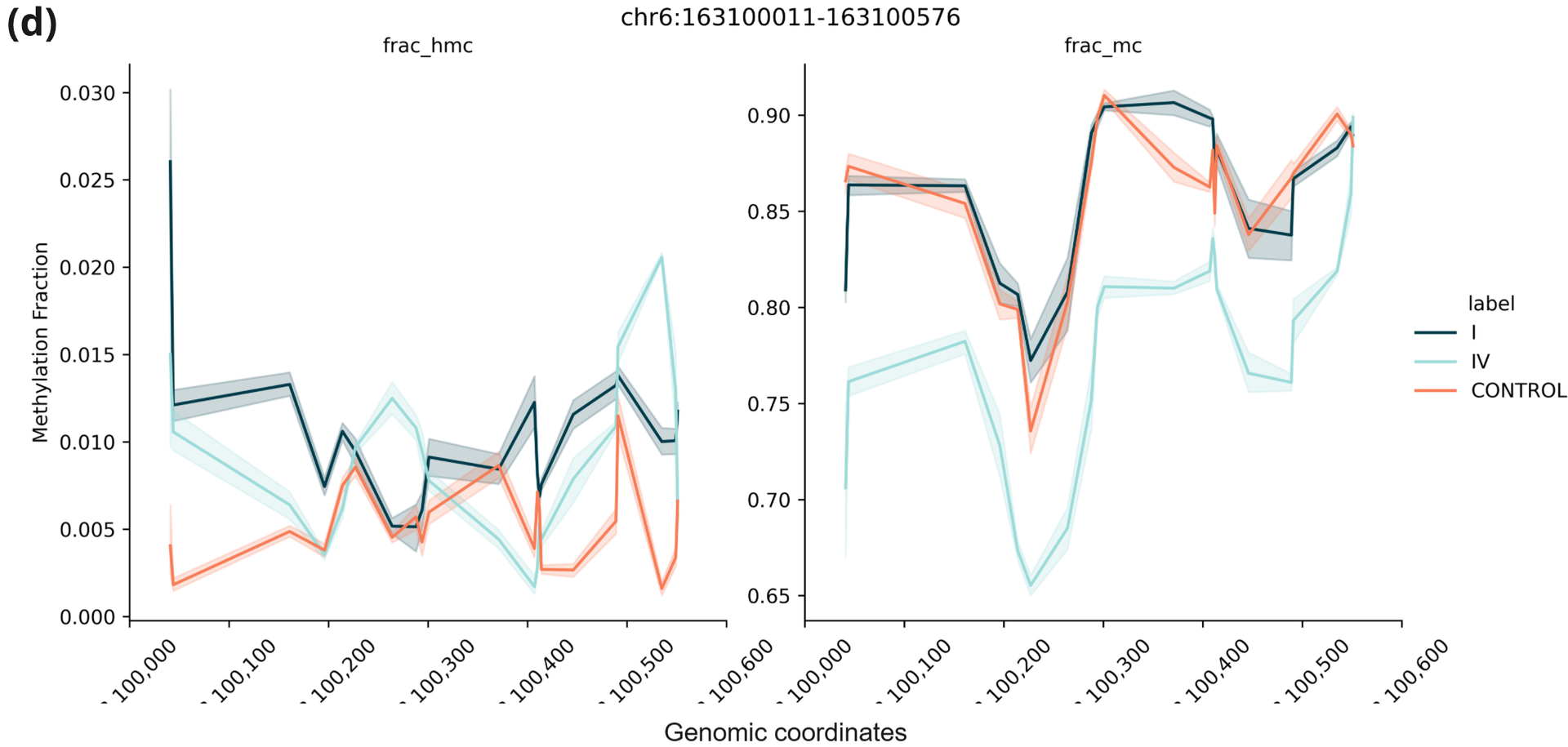
Figure 3.
- Schematic of the analysis of 6-base data from cfDNA focussing on 5mC and 5hmC in regions that are derived from DMRs between CRC tissue vs adjacent normal tissue.
- Volcano plot for DMRs between stage IV CRC tissue and adjacent matched normal, using data from TCGA². Colour indicates density of points, with yellow used to indicate a higher density.
- Venn diagram of tissue-derived DMRs where there are also statistically significant (q < 0.05) differences in 5mC or 5hmC when analysing stage I CRC vs healthy cfDNA.
- Trace plots of 5hmC (left) and 5mC (right) fractions in an example region that showed increased 5hmC in stage I plasma and decreased 5mC in stage IV plasma.
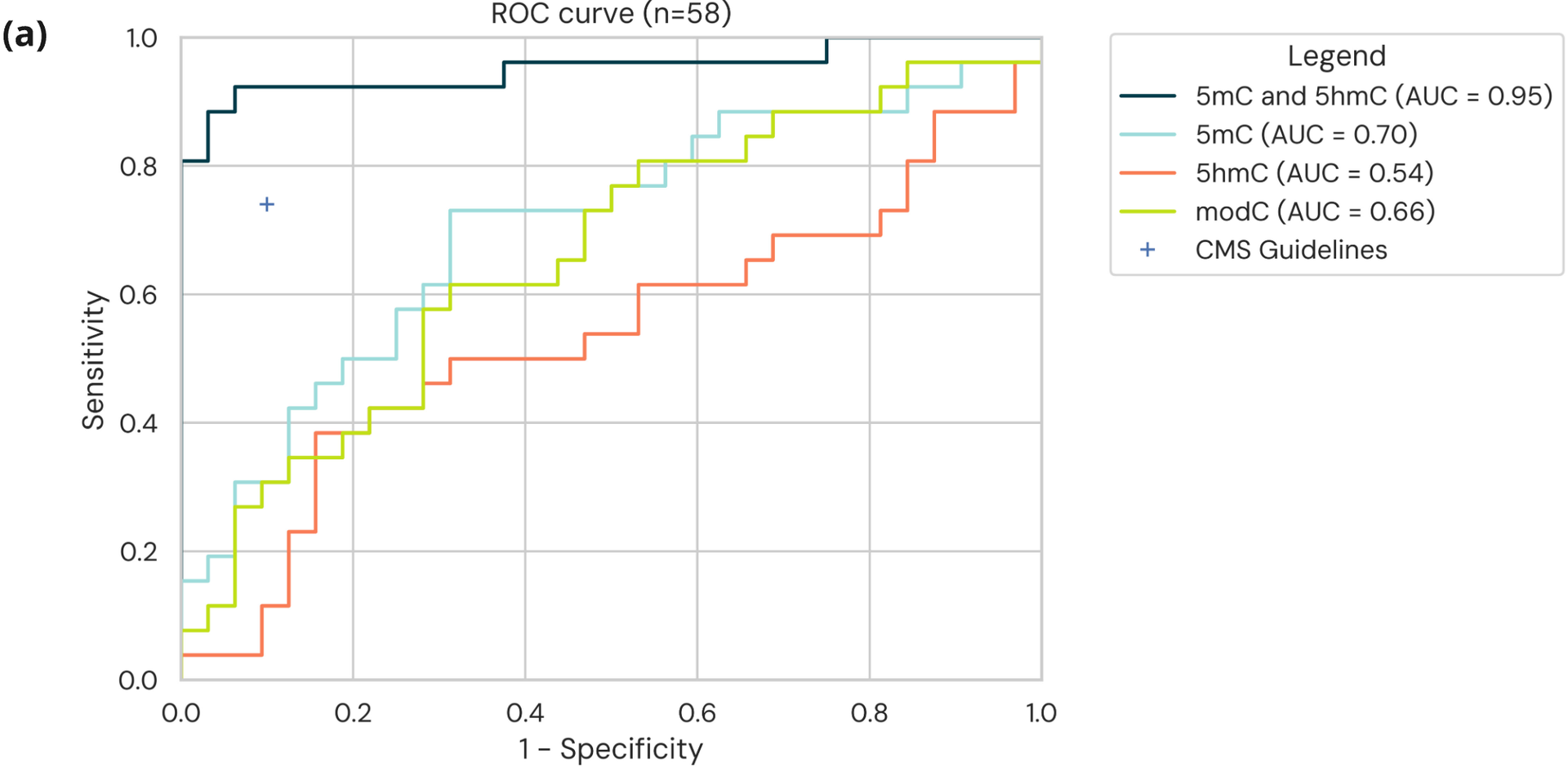
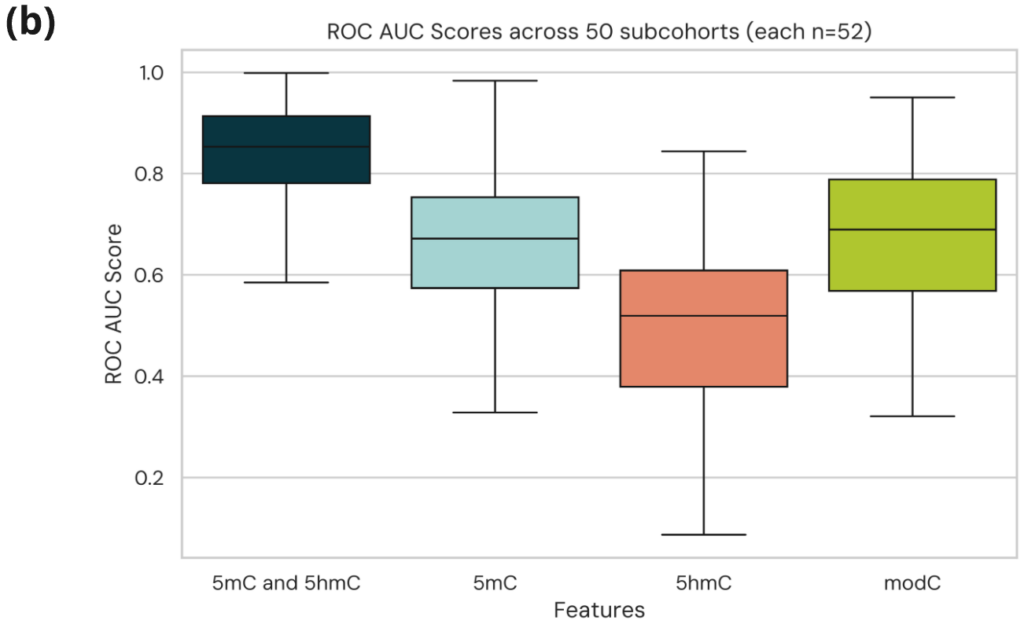
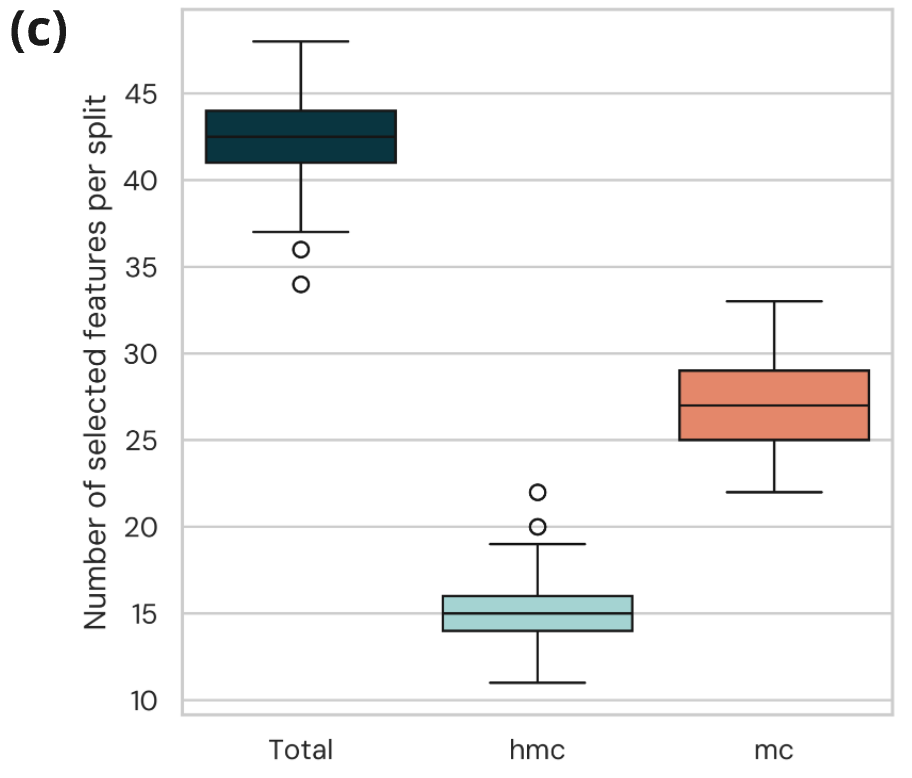
Figure 4.
- ROC curves for modC only, 5mC only, 5hmC only, and 5mC and 5hmC models using generalised linear models and a leave-one-out cross validation approach³ ⁴ show the combination of 5mC and 5hmC is better for detection of stage I CRC from cfDNA than 5mC, 5hmC or 5modC alone.
- Box plot showing AUCs across 500 sub-cohorts for the different models demonstrate 5mC and 5hmC are consistently the most performative and the discriminatory signal is robust to cohort changes.
- Classifiers consistently select a mixture of 5mC and 5hmC features across 58 cross-validation splits, further evidencing the synergism between 5mC and 5hmC for the early detection of CRC.
This study shows that 5mC and 5hmC taken together are highly effective as biomarkers for early-stage CRC detection in cfDNA. The combination of these biomarkers shows a clear advantage over the use of either either individually as well as a significant improvement over the use of the conflated modC biomarkers and clearly demonstrates that 5hmC is important for the detection of changes that occur early in disease course. We feel there is merit in applying duet evoC 6-base sequencing to larger clinical cohorts, across different diseases to more broadly evaluate the potential of 6-base data to improve early disease detection.
- Füllgrabe J. et al. Simultaneous sequencing of genetic and epigenetic bases in DNA. Nat Biotechnol. 2023 Oct;41(10):1457-1464.
- https://portal.gdc.cancer.gov/, January 2024
- Friedman J. et al.. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1-22.
- Tay J.K. et al. Elastic Net Regularization Paths for All Generalized Linear Models. J Stat Softw. 2023;106:1.