- Fabio Puddu,
- Tom Charlesworth
- Robert Crawford
- Nick Harding
- Annelie Johansson
- Ermira Leslie
- Casper K Lumby
- Aurelie Modat
- Jamie Scotcher
- Michael S Wilson
- Shirong Yu
- Jens Füllgrabe
- Walraj S Gosal
- Páidí Creed
Liquid biopsy for profiling of cell free DNA (cfDNA) in blood holds huge promise to transform how we experience and manage cancer by early detection and identification of residual disease and subtype. However, a standard blood draw yields an average of only 10 ng of cfDNA, of which DNA derived from the tumour is a small minority.
Genetic and methylation data together have been shown to be more powerful for the detection of early cancer than either alone. Constrained to measuring four states of information, existing NGS-based technologies sacrifice genetic information for methylation calling.
| State | Standard sequencing protocol | Protocol with C→T deamination |
|---|---|---|
| 1 | A | A |
| 2 | c/mC/hmC | mC/hmC |
| 3 | G | G |
| 4 | T | C/T |
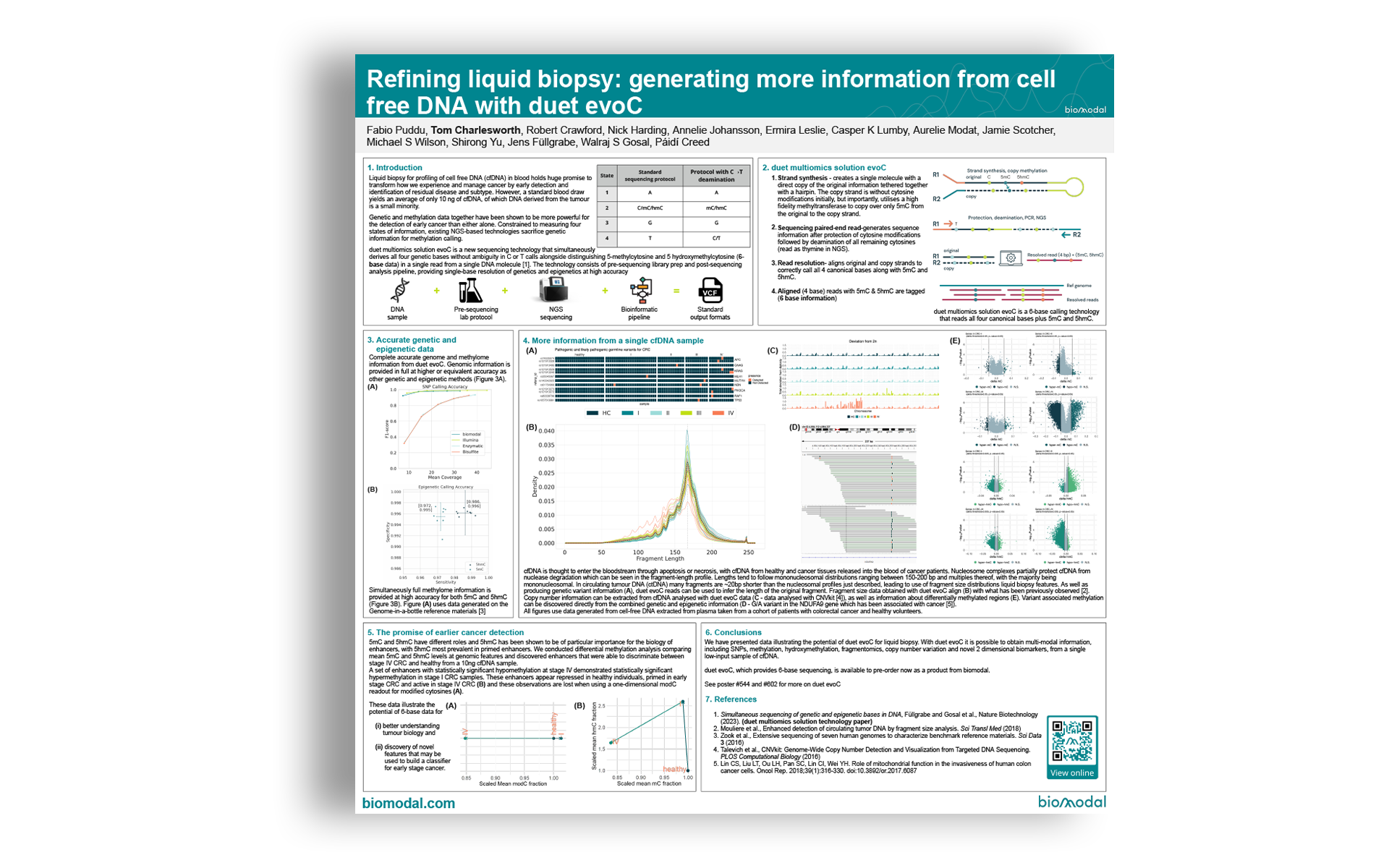
duet multiomics solution evoC is a new sequencing technology that simultaneously derives all four genetic bases without ambiguity in C or T calls alongside distinguishing 5-methylcytosine and 5 hydroxymethylcytosine (6-base data) in a single read from a single DNA molecule. The technology consists of pre-sequencing library prep and post-sequencing analysis pipeline, providing single-base resolution of genetics and epigenetics at high accuracy

- Strand synthesis – creates a single molecule with a direct copy of the original information tethered together with a hairpin. The copy strand is without cytosine modifications initially, but importantly, utilises a high fidelity methyltransferase to copy over only 5mC from the original to the copy strand.
- Sequencing paired-end read-generates sequence information after protection of cytosine modifications followed by deamination of all remaining cytosines (read as thymine in NGS).
- Read resolution– aligns original and copy strands to correctly call all 4 canonical bases along with 5mC and 5hmC.
- Aligned (4 base) reads with 5mC & 5hmC are tagged (6 base information)
duet multiomics solution evoC is a 6-base calling technology that reads all four canonical bases plus 5mC and 5hmC.
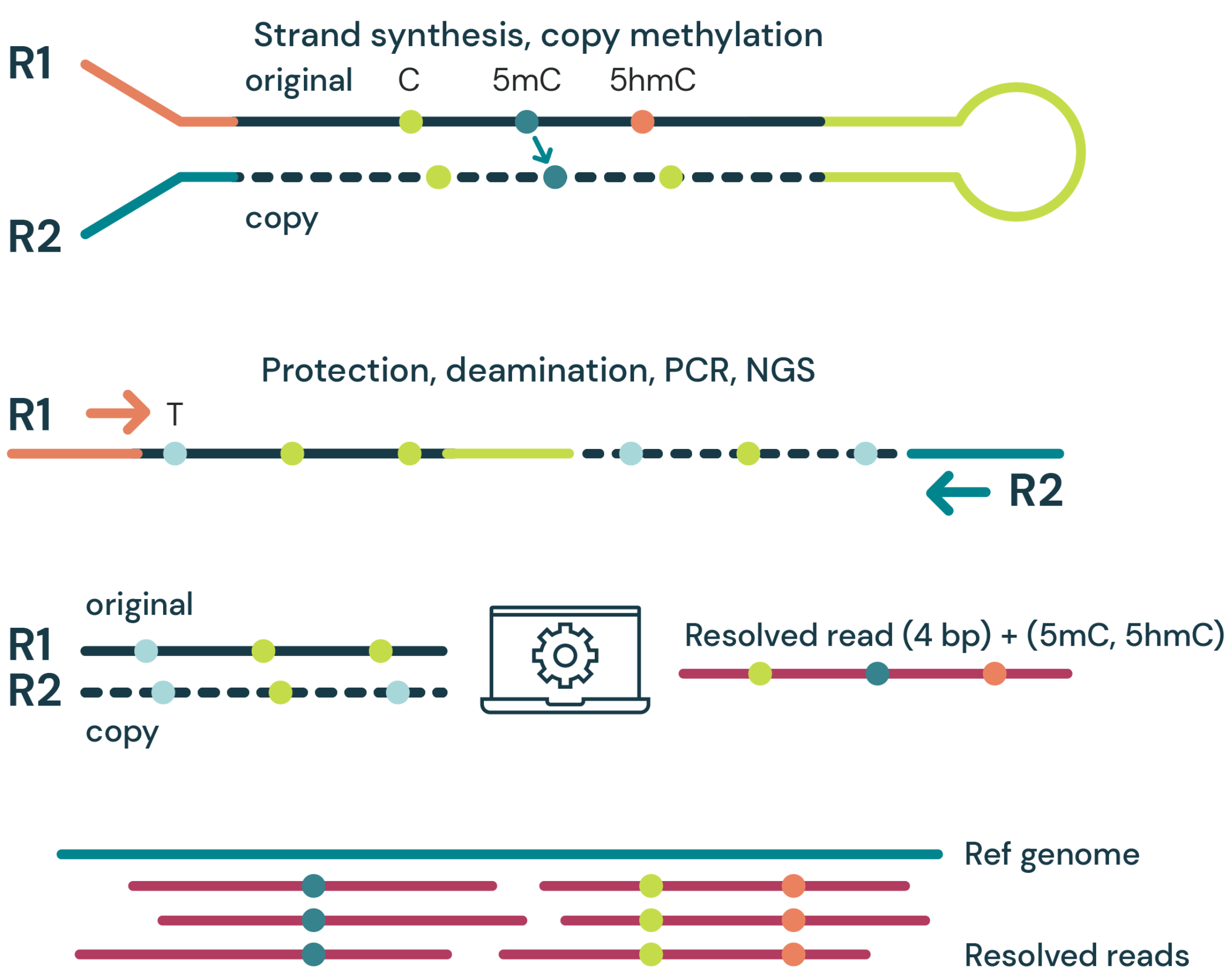
Complete accurate genome and methylome information from duet evoC. Genomic information is provided in full at higher or equivalent accuracy as other genetic and epigenetic methods (Figure 3A).
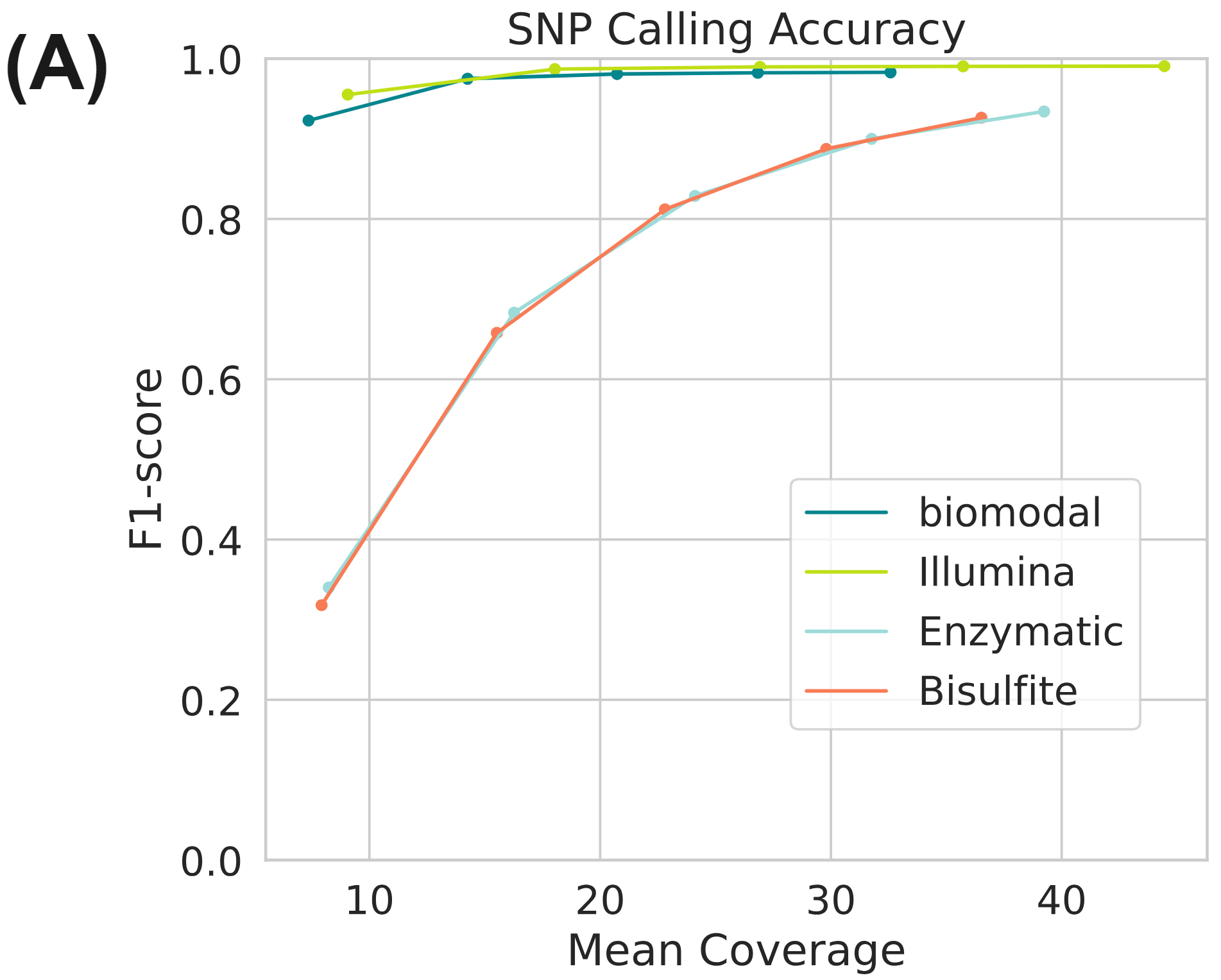
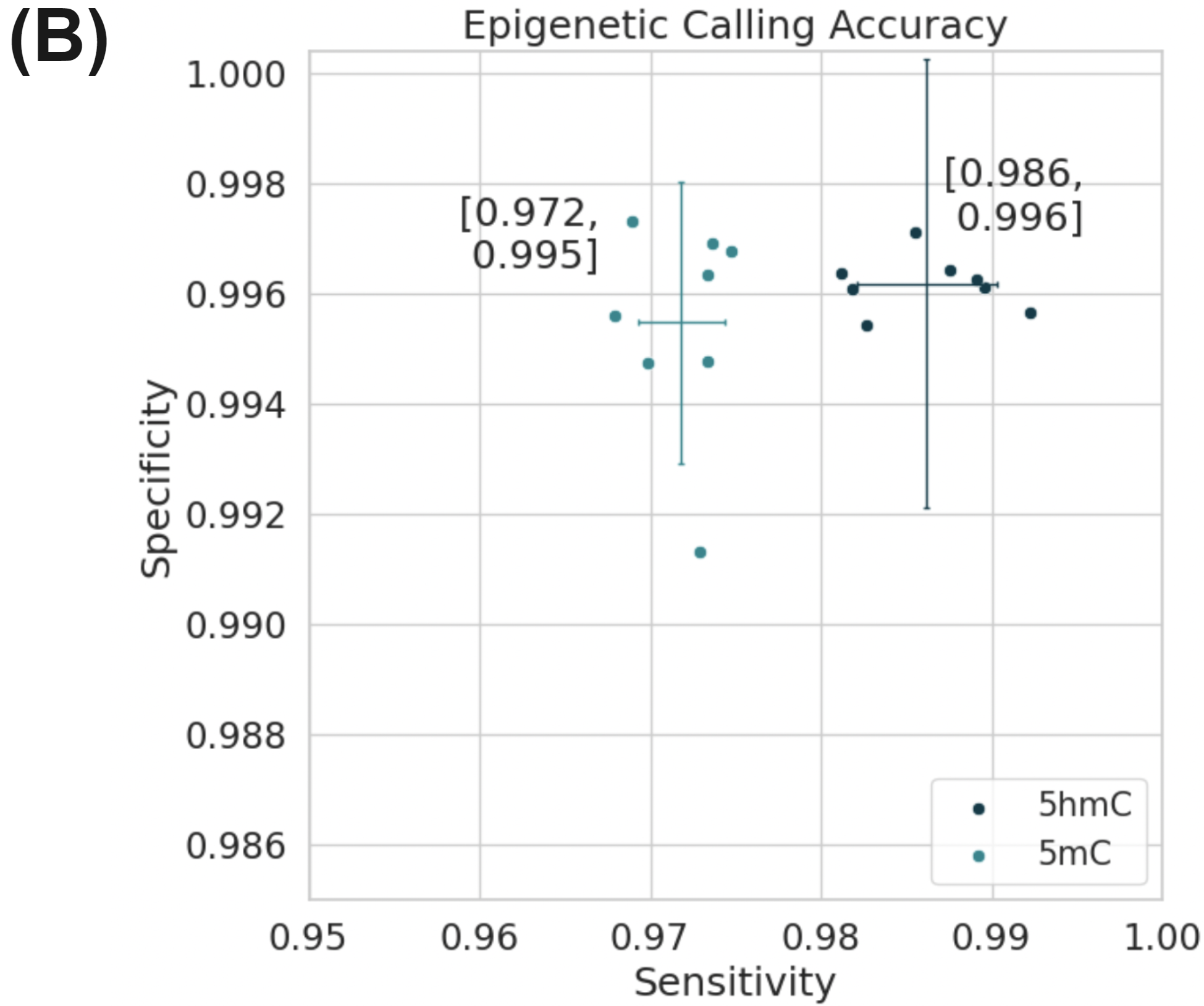
Simultaneously full methylome information is provided at high accuracy for both 5mC and 5hmC (Figure 3B). Figure (A) uses data generated on the Genome-in-a-bottle reference materials.
Complete accurate genome and methylome information from duet evoC. Genomic information is provided in full at higher or equivalent accuracy as other genetic and epigenetic methods (Figure 3A).
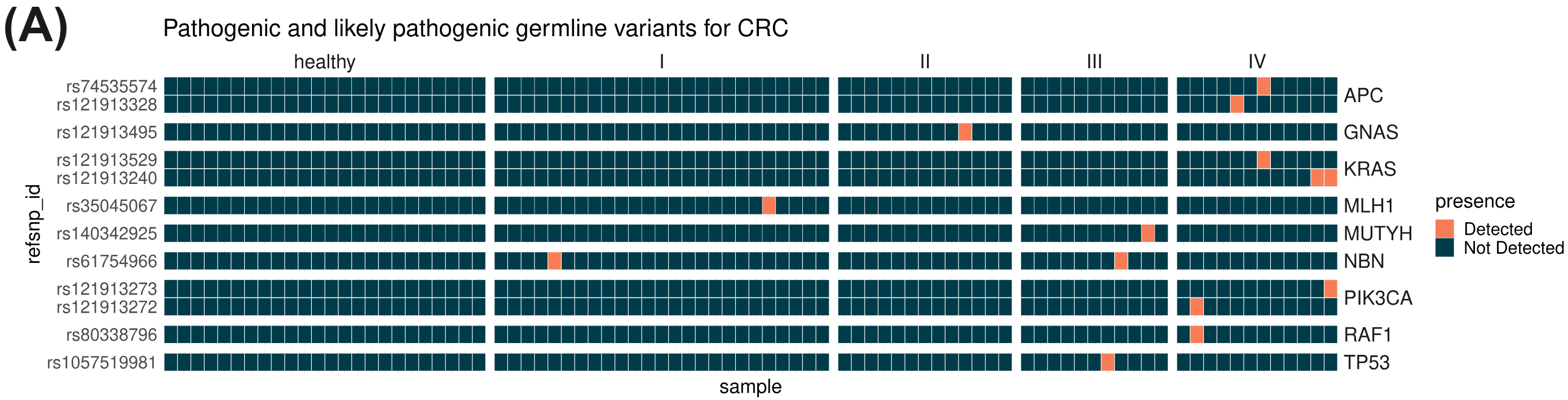
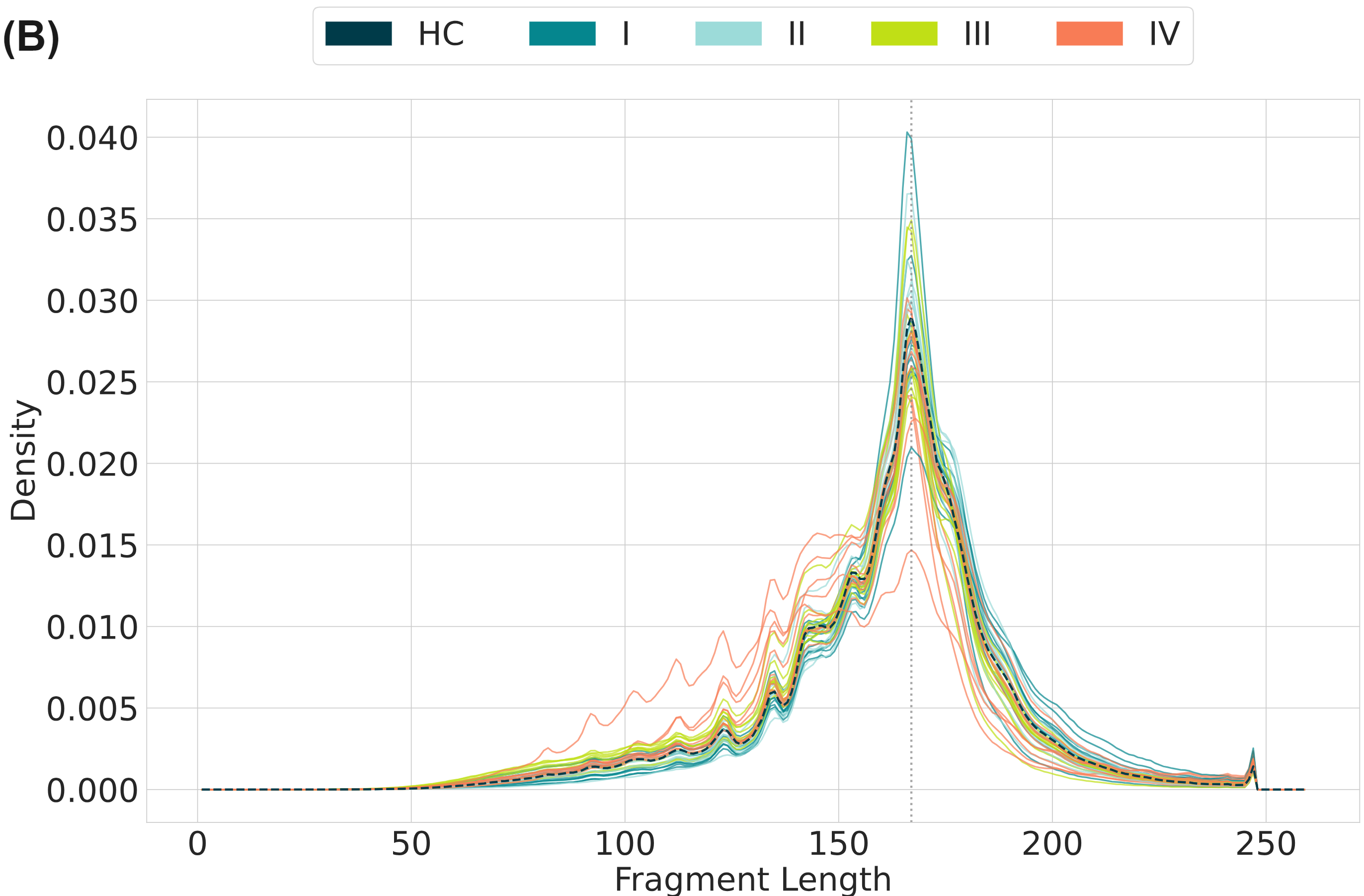
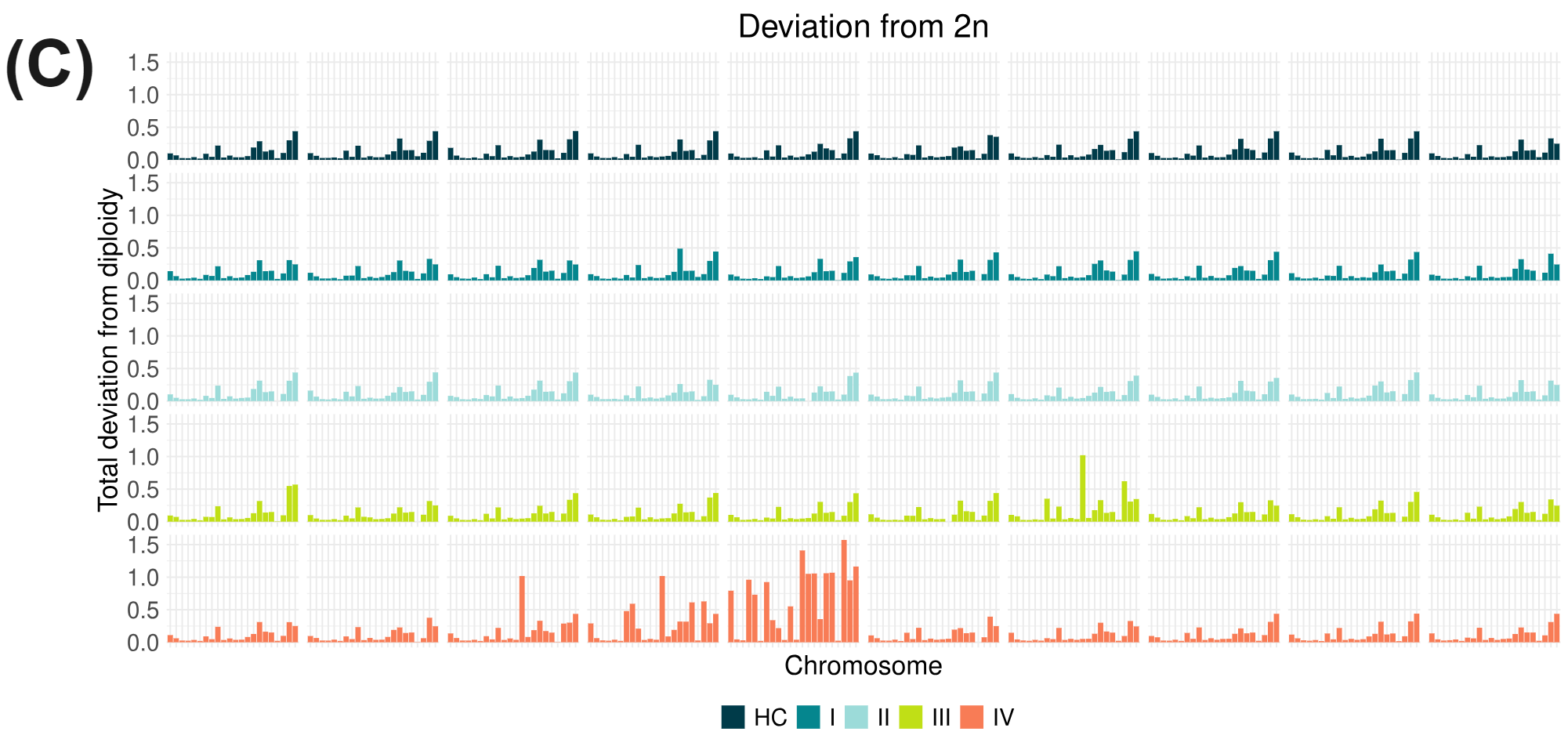
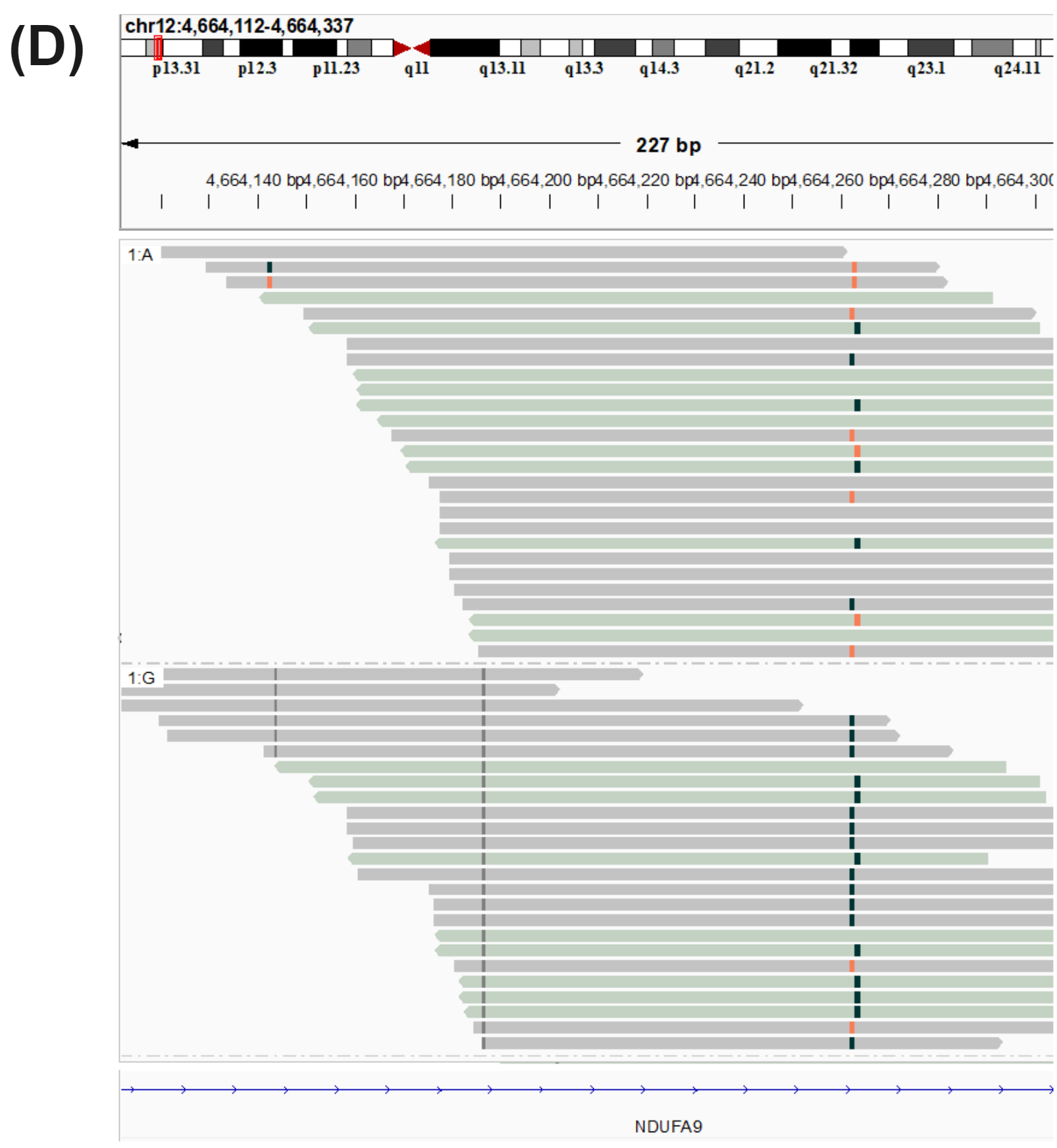
cfDNA is thought to enter the bloodstream through apoptosis or necrosis, with cfDNA from healthy and cancer tissues released into the blood of cancer patients. Nucleosome complexes partially protect cfDNA from nuclease degradation which can be seen in the fragment-length profile. Lengths tend to follow mononucleosomal distributions ranging between 150-200 bp and multiples thereof, with the majority being mononucleosomal. In circulating tumour DNA (ctDNA) many fragments are ~20bp shorter than the nucleosomal profiles just described, leading to use of fragment size distributions liquid biopsy features. As well as producing genetic variant information (A), duet evoC reads can be used to infer the length of the original fragment. Fragment size data obtained with duet evoC align (B) with what has been previously observed. Copy number information can be extracted from cfDNA analysed with duet evoC data (C – data analysed with CNVkit), as well as information about differentially methylated regions (E). Variant associated methylation can be discovered directly from the combined genetic and epigenetic information (D – G/A variant in the NDUFA9 gene which has been associated with cancer).
All figures use data generated from cell-free DNA extracted from plasma taken from a cohort of patients with colorectal cancer and healthy volunteers.
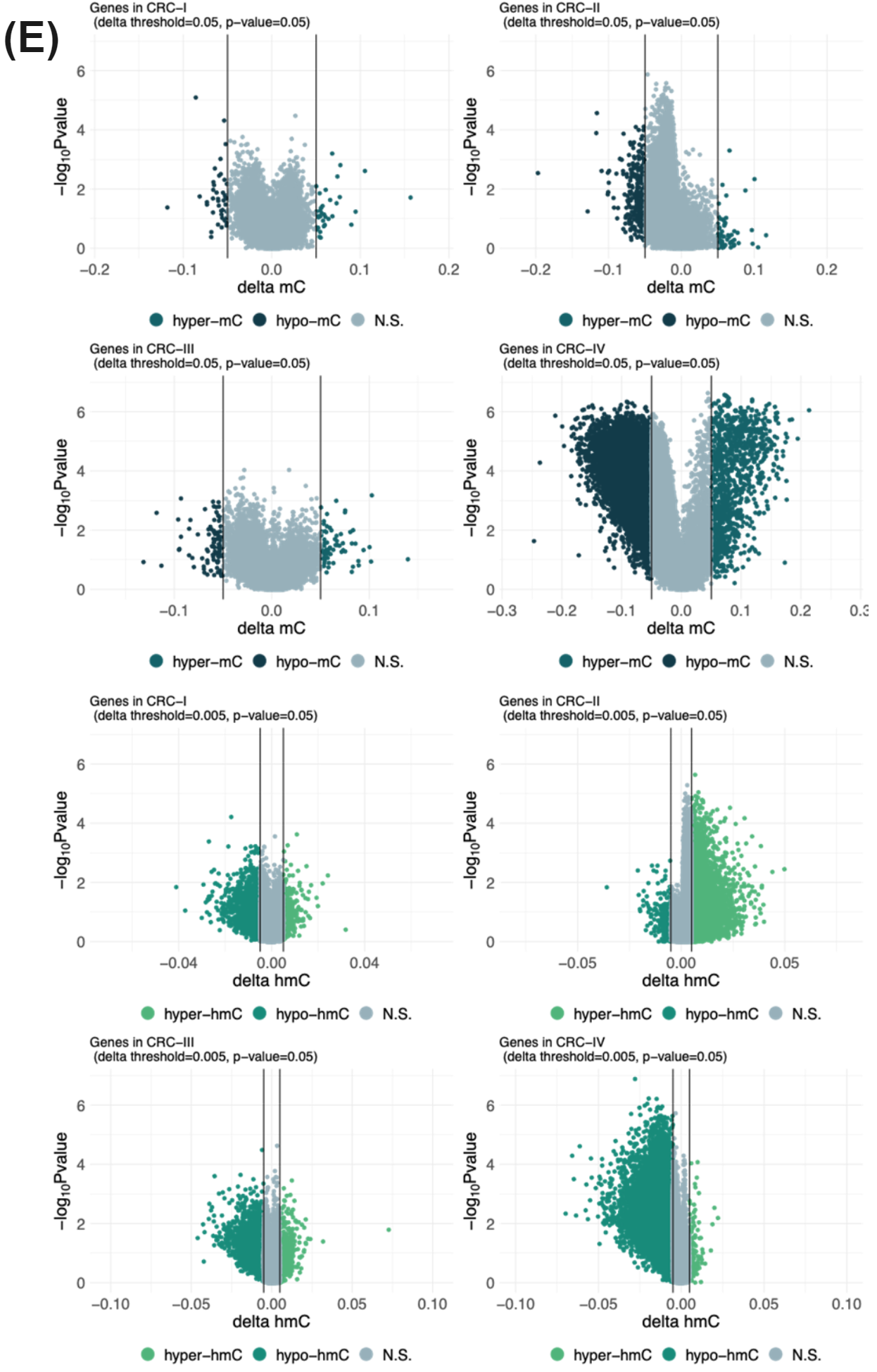
5mC and 5hmC have different roles and 5hmC has been shown to be of particular importance for the biology of enhancers, with 5hmC most prevalent in primed enhancers. We conducted differential methylation analysis comparing mean 5mC and 5hmC levels at genomic features and discovered enhancers that were able to discriminate between stage IV CRC and healthy from a 10ng cfDNA sample.
A set of enhancers with statistically significant hypomethylation at stage IV demonstrated statistically significant hypermethylation in stage I CRC samples. These enhancers appear repressed in healthy individuals, primed in early stage CRC and active in stage IV CRC (B) and these observations are lost when using a one-dimensional modC readout for modified cytosines (A).
These data illustrate the potential of 6-base data for (i) better understanding tumour biology and (ii) discovery of novel features that may be used to build a classifier for early stage cancer.
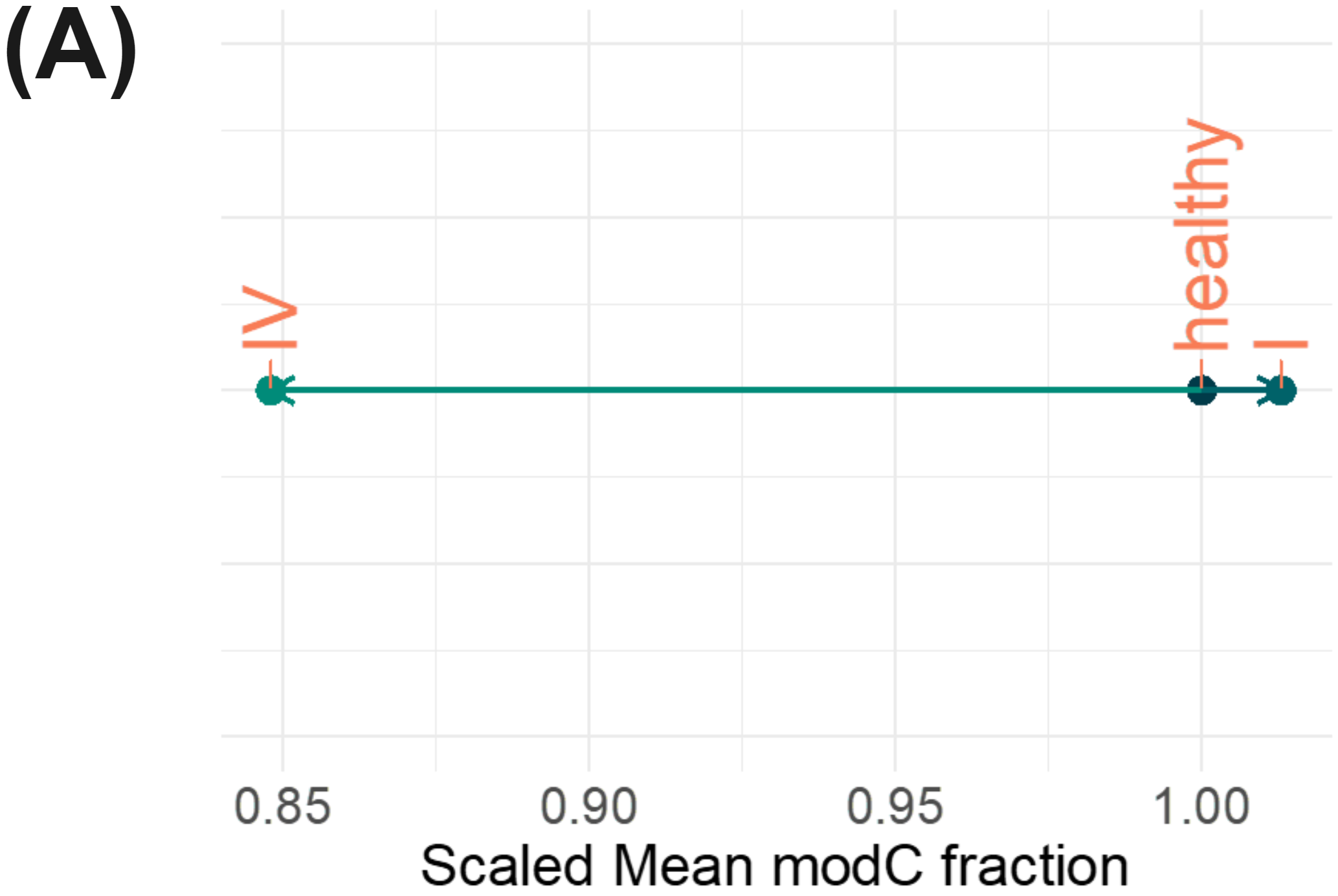
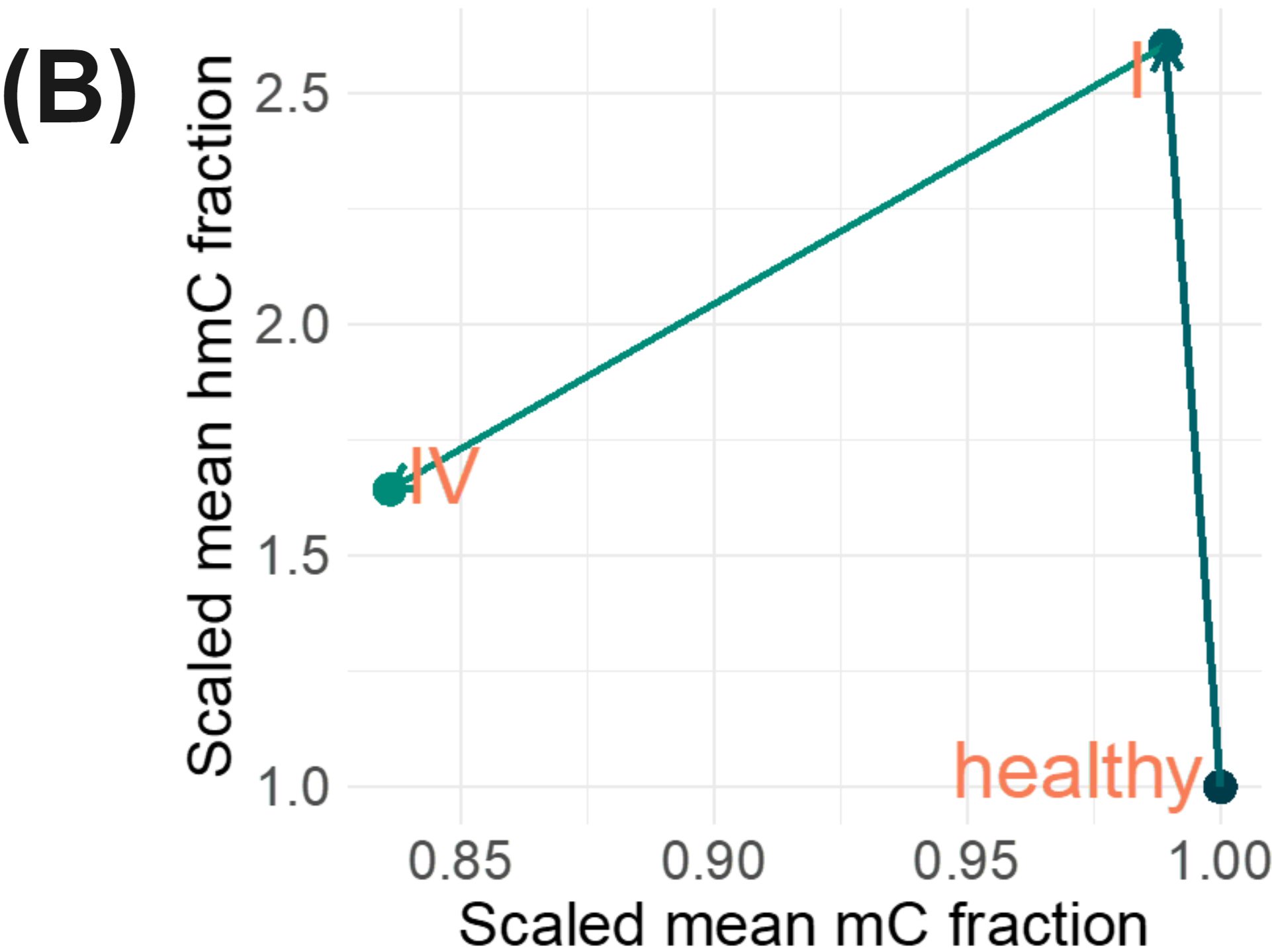
We have presented data illustrating the potential of duet evoC for liquid biopsy. With duet evoC it is possible to obtain multi-modal information, including SNPs, methylation, hydroxymethylation, fragmentomics, copy number variation and novel 2 dimensional biomarkers, from a single low-input sample of cfDNA.
See these posters for more on duet evoC: