Learn how simultaneous measurement of genetics and epigenetics with duet multiomics solution +modC enables the study of allele-specific methylation and how this can be used to understand the impact of epigenetic rejuvenation.
biomodal
- Nicholas J Harding
- Páidí Creed
- David Currie
- Sabri Jamal
- Casper K Lumby
- Jack M Monahan
- David J Morley
- Fabio Puddu
- Jamie Scotcher
- Rosie Telford Spencer
- Jean Teyssandier
- Michael Wilson
- Jens Fullgrabe
- Audrey Vandomme
- Aurel Negrea
- Alexandra Palmer
- Philippa Burns
- Shirong Yu
- Joanna D Holbrook
Altos Labs & Babraham Institute
- Diljeet Gill
- Aled Parry
- Wolf Reik
There is more to DNA than A, C, G and T. Epigenetics plays a causal role in cell fate, ageing and disease development. Methylated cytosines, such as 5mC and 5hmC, represent important biomarkers and are informally considered the 5th and 6th letters in the genetic alphabet.
Genetic and methylation data together on the same read allow us to get a more complete picture of genome biology. Constrained to measuring four states of information, existing NGS-based technologies sacrifice genetic for methylation calling.
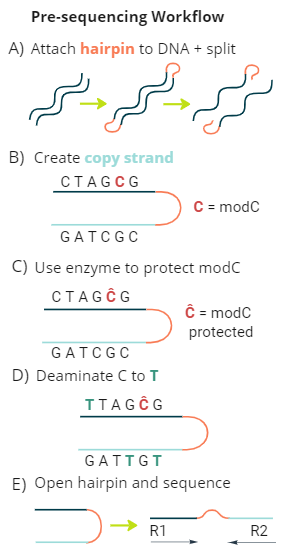
duet multiomics solution +modC is a new sequencing technology that derives all four genetic bases without ambiguity in C or T calls, plus epigenetic modified cytosine (modC.) The technology consists of pre-sequencing workflow and post-sequencing analysis pipeline, providing single-base resolution of genetics and epigenetics at high accuracy.
|
State
|
Standard
sequencing protocol
|
Protocol with C→T deamination
|
|
1
|
A
|
A
|
|
2
|
C/mC/hmC
|
mC/hmC
|
|
3
|
G
|
G
|
|
4
|
T
|
C/T
|

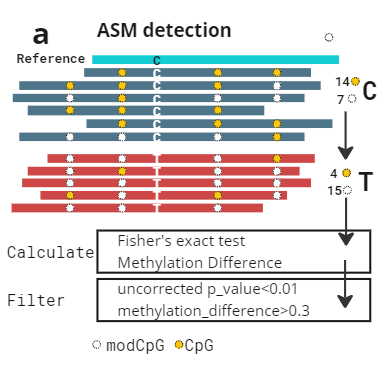
Having both genetic and epigenetic information on the same read allows us to assign reads to haplotypes with high accuracy. We can therefore identify regions of allele specific methylation without many of the assumptions of models that rely on deamination based methods. Reads are assigned to haplotypes based on observing either allele of a heterozygote (called by GATK HaplotypeCaller).
left: Schematic of ASM detection. Reads are assigned to haplotypes: then CpG/modCpG are counted. A statistical test determines the p-value associated with the alternative hypothesis of ASM in this region. Note that in deamination based methods there is extremely limited ability to detect C > T mutations.
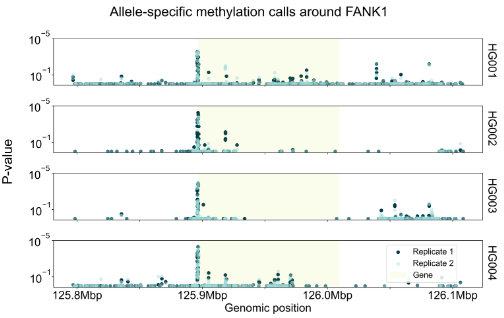
We have created a benchmark dataset, using all 7 Genome in a Bottle samples (2x technical replicates per sample). We called ASM on all samples using the above method. We observe promoter regions are enriched for ASM calls, and many genes are consistently highlighted across multiple samples, e.g. FANK1.
right: Figure showing ASM calls in one of the genes consistently associated with ASM across samples. y-axis shows p value associated with each heterozygote. Samples HG005-7 not shown due to space but display same peak.
We validated our ASM calls on the above data using external datasets. We show imprinted regions (defined by Jima et al.) are highly enriched for ASM calls. Additionally given X-silencing is mediated by methylation, we further validate our calls against published X-inactivated genes (Kravitz et al).
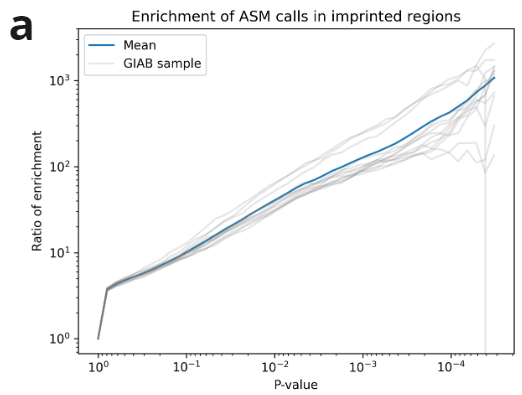
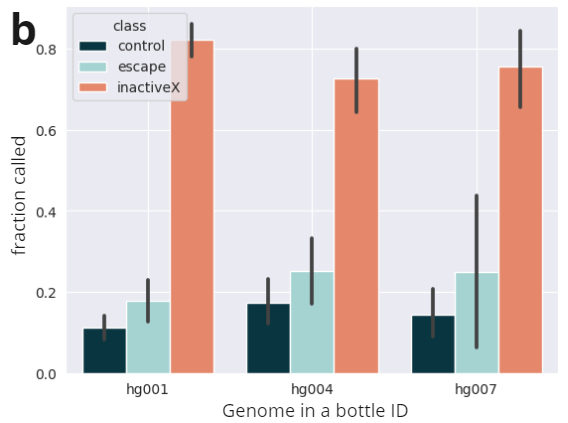
- ASM show high enrichment of heterozygotes in imprinted regions, increasing with p-value.
- X-inactivated genes are consistently identified as silenced across samples and studies. Escape genes are present on the X, but are not consistently silenced. Controls are randomly selected from autosomes (Kravitz et al). We observe enriched calls in X-inactivated genes in female samples.
At the cellular level, ageing is associated with reduced function, altered gene expression and a perturbed epigenome (Horvath 2013). Recent work has demonstrated that the epigenome and some elements of function are rejuvenated by the maturation phase of iPSC reprogramming, without loss of cell identity.
Here we apply the Gill et al. (2022) “maturation phase transient reprogramming” (MPTR), where reprogramming factors (OSKM) are expressed until the rejuvenation point and then withdrawn. We use the novel +modC technology to investigate genetic and epigenetic changes after transient reprogramming.
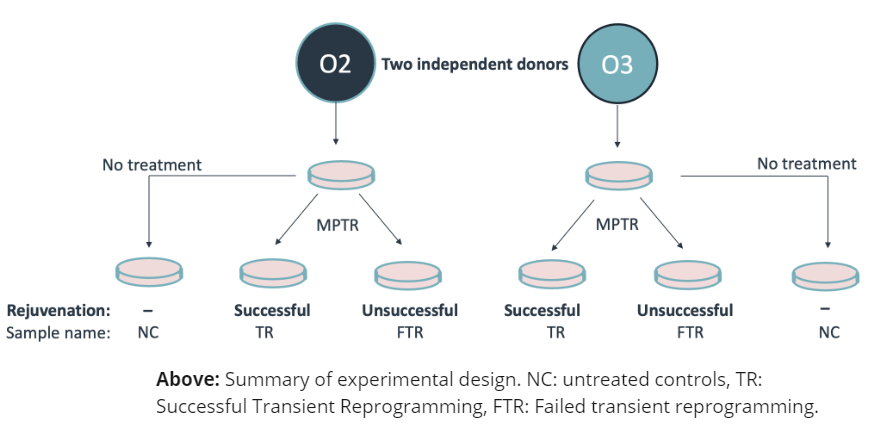
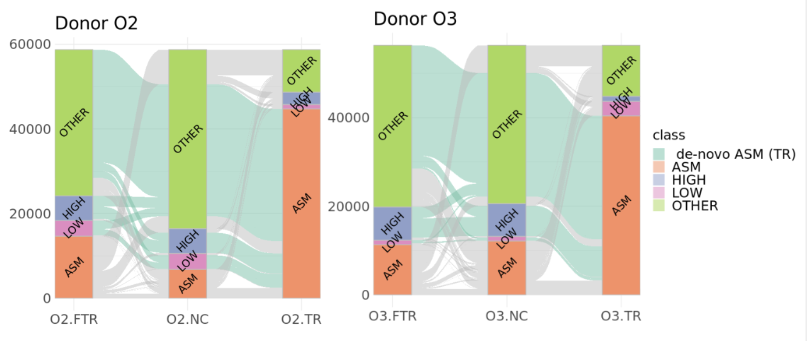
The heterozygous sites gaining ASM in TR samples are largely associated with intermediate methylation in NC samples (“OTHER”) or to a smaller extent with fully methylated (“HIGH”) or demethylated (“LOW”) sites. The majority of these sites are not converted to ASM in FTR samples.
right: Sankey representation of methylation status changes between untreated controls (NC), successfully rejuvenated cells (TR), and cells that failed rejuvenation (FTR). Successful MPTR is associated with increased ASM in both donors.
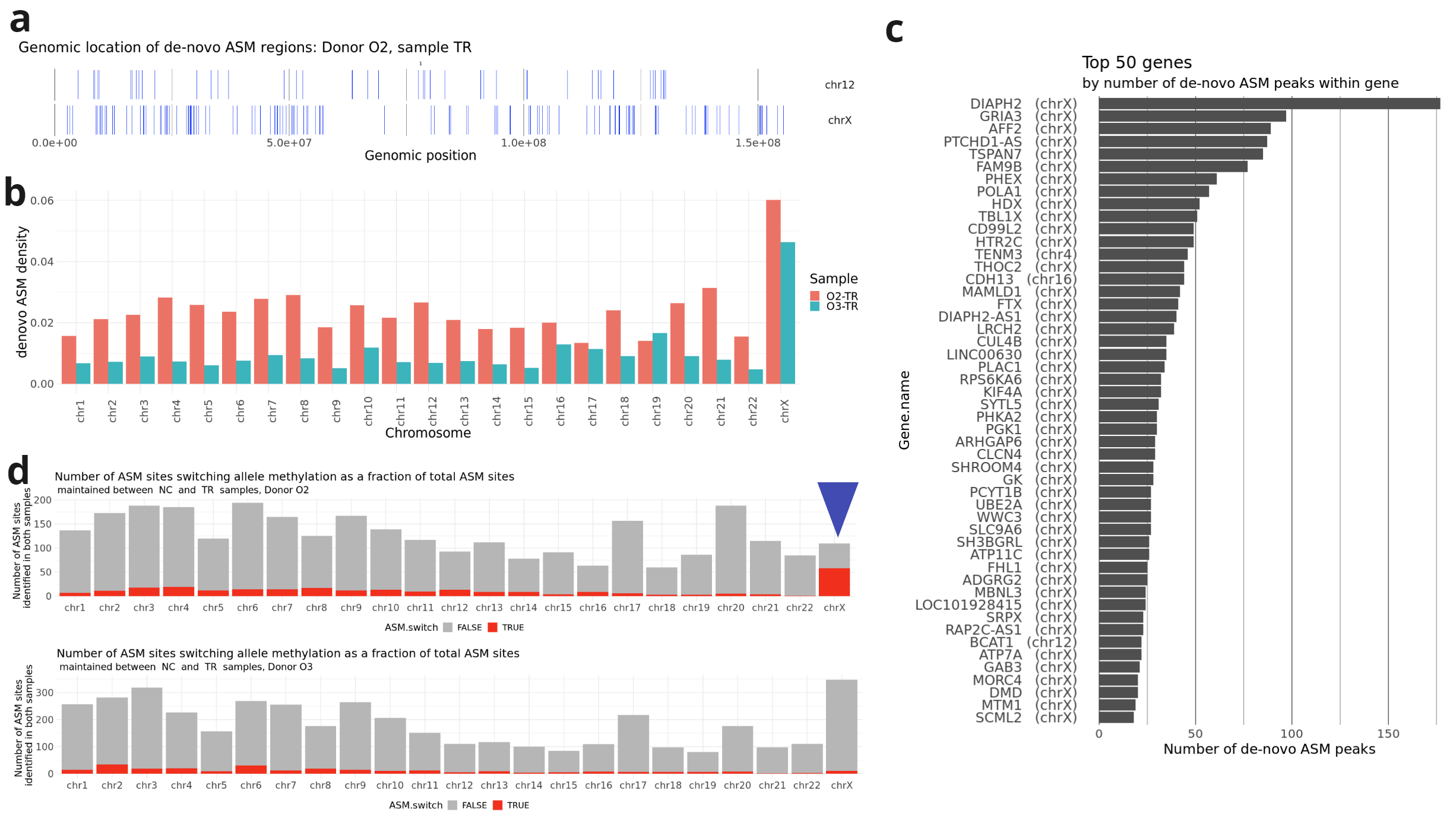
- Localisation of genomic windows (vertical blue lines) showing de-novo ASM in TR samples on chromosome X and a chromosome of comparable length (chr12) for donor O2;
- Density of de novo ASM in TR samples from different donors, calculated as the length of windows displaying de novo ASM divided by the total length of the chromosome.
- Top 50 genes by number of de novo ASM windows within the gene body in both O2 and O3 donors. Most genes are located on chromosome X.
- In donor O2 (but not in donor O3), approximately half of the ASM sites that retained their ASM status during MPTR switched the methylation from one allele to the other.
- Simultaneous sequencing of genetic and epigenetic bases in DNA. Füllgrabe and Gosal et al., Nature Biotechnology (2023). (duet multiomics solution technology paper)
- Extensive sequencing of seven human genomes to characterize benchmark reference materials. Zook et al. Scientific Data (2016).
- Genomic map of candidate human imprint control regions: the imprintome. Jima et al. Epigenetics. (2022)
- Random allelic expression in the adult human body. Kravitz et al. Cell Reports (2023)
- Multi-omic rejuvenation of human cells by maturation phase transient reprogramming. Gill et al., eLife (2022). Apr 8;11:e71624.
- DNA methylation age of human tissues and cell types. Horvath et al, Genome Biology (2013);14(10):R115.