Get multimodal insights into the complexity of ageing
Epigenetic changes including DNA methylation contribute to the complex process of ageing by influencing gene expression patterns and cellular function, affecting various biological pathways such as cell repair, metabolism, and the immune response. Epigenetic changes can be influenced by several factors, including response to environment, lifestyle, and exposure to disease. Alterations in methylation patterns can lead to age-related changes in cellular function and likely contribute to pathologies such as cancer, arthritis, and cardiovascular and neurodegenerative diseases.
DNA methylation and changes in epigenetic regulatory systems have been directly correlated with longevity in many organisms, including humans, and can provide insights into biological age and the potential for interventions to delay ageing and extend healthy lifespan.
Ageing refers to the natural, progressive process of physiological and cellular changes over time. But the rate of ageing differs between individuals – as an individual ages, their epigenetic profile will change in response to their environment and lifestyle, leading to shifts in cellular function and homeostasis.
Chronological age
This is the simplest measure of age, based simply on time since birth. Chronological age is absolute and uniform for everyone, so two people born on the same day will have the same chronological age, regardless of their health, lifestyle, or environmental factors.
Biological age
Reflects the functional state of your cells, tissues, and organs compared to your actual chronological age. Biological age is influenced by factors such as genetics, environment, and lifestyle and so provides a more accurate assessment of an individual's overall health and ageing trajectory compared to chronological age.
Levels of specific genes are used as epigenetic clocks, developed through the analysis of large sets of methylation data across multiple genes from a diverse population. Statistical and machine learning methods are then used to identify specific sets of CpG sites whose levels of 5mC correlate strongly with chronological age. These methylation profiles are then used to create predictive models that can estimate the biological age of a tissue, cell, or whole organism.
The interplay between 5mC and 5hmC is crucial for the dynamic regulation of gene expression and maintenance of cellular and genomic integrity during ageing. Alterations in the balance between methylation and demethylation, as represented by changes in the levels and distribution of 5mC and 5hmC, can lead to age-related physiological changes and increase the risk of diseases. Their roles in cellular processes are central to understanding the molecular mechanisms of ageing and identifying potential biomarkers for disease.
Gene regulation
High levels of 5mC at gene promoters are associated with gene silencing. Changes in 5mC patterns with age can lead to the inappropriate expression or silencing of genes critical for maintaining cellular function, contributing to the ageing process and the development of age-related diseases.
Zemach A. McDaniel IE. Silva P. Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919.
Genome stability
Maintaining genome integrity is crucial for cellular longevity and function. 5mC helps suppress transposable elements and maintain chromosome structure, thereby reducing genomic instability that is associated with ageing.
Gonzalo S. Jaco I. Fraga MF. Chen T. Li E. Esteller M. Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424.
Li E. Bestor TH. Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926.
Cellular senescence
The accumulation of senescent cells contributes to ageing and age-related pathologies. The distribution of 5mC changes in senescent cells, affecting the expression of senescence-associated genes and inflammatory factors.
Zhang W. Ji W. Yang J. Yang L. Chen W. Zhuang Z. Comparison of global DNA methylation profiles in replicative versus premature senescence. Life Sci. 2008;83:475–480.
So AY. Jung JW. Lee S. Kim HS. Kang KS. DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS One. 2011;6:e19503.
Gene expression and neurogenesis
Studies have shown that changes in 5hmC are associated with psychiatric disorders and several neurodegenerative disease, which can also be age-related.
Klein CJ. Botuyan MV. Wu Y. Ward CJ. Nicholson GA. Hammans S. Hojo K. Yamanishi H. Karpf AR. Wallace DC. Simon M. Lander C. Boardman LA. Cunningham JM. Smith GE. Litchy WJ. Boes B. Atkinson EJ. Middha S. B Dyck PJ. Parisi JE. Mer G. Smith DI. Dyck PJ. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600.
Active DNA demethylation
5hmC is an intermediate in the process of active DNA demethylation, which can activate gene expression. This dynamic regulation of gene expression is crucial for responding to environmental changes and stressors throughout ageing.
Fraga MF. Ballestar E. Paz MF. Ropero S. Setien F. Ballestar ML. Heine-Suñer D. Cigudosa JC. Urioste M. Benitez J. Boix-Chornet M. Sanchez-Aguilera A. Ling C. Carlsson E. Poulsen P. Vaag A. Stephan Z. Spector TD. Wu Y-Z. Plass C. Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA.
Genome stability and repair
5hmC may also play a role in DNA repair mechanisms and maintaining genome stability, which are vital for preventing age-related genomic damage and cellular decline.
Dubec SJ. Aurora R. Zassenhaus HP. Mitochondrial DNA mutations may contribute to aging via cell death caused by peptides that induce cytochrome c release. Rejuvenation Res. 2008;11:611–619.
Ageing is accompanied by changes in genetic sequence and DNA methylation so a full understanding of both genetic and epigenetic information is vital to study the effects of ageing. Mapping genetic variants and methylation status of 5mC and 5hmC simultaneously on the same DNA molecule can provide valuable insight into how the two systems interact in the ageing cell.
- Epigenomic and genomic information on the same DNA fragment from a single low-input sample in one workflow
- Distinguish 5mC and 5hmC and variant-associated methylation without data loss
- High sensitivity and specificity for SNP detection especially C-to-T mutations
- More information from a single workflow with a low sample input requirement (5ng DNA)
The term variant-associated methylation (VAM) can be used to describe the direct association of genetic variation with changes in DNA methylation – either in close proximity or across large genomic distances. VAM directly links genetic sequence information with 5mC and 5hmC methylation data to better understand the dynamic interactions that are occurring in a cell.
The duet multiomics solution evoC gives combined genetic and epigenetic sequence data and access to the 6-base genome to uncover the cellular processes that govern ageing. All 4 canonical bases and distinguish 5mC and 5hmC from a single 5ng DNA sample, in a single experiment, with high accuracy.
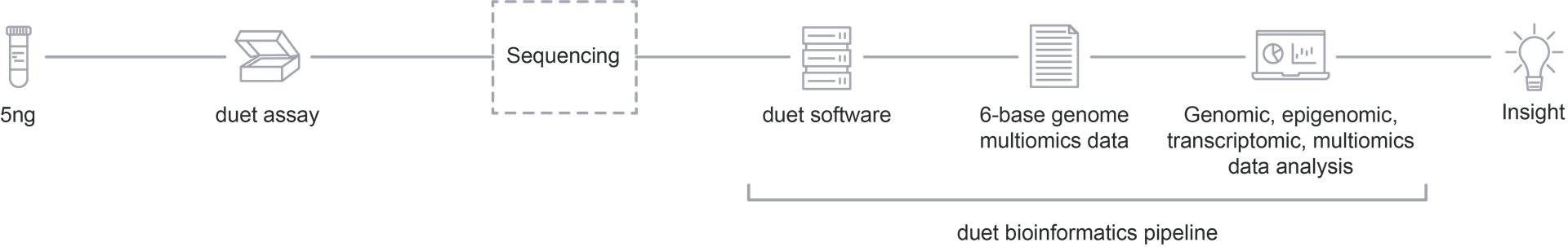
We can help you reveal new data and multimodal insights from your research.