Join biomodal at this two-day in-person conference at Harvard Medical School. Day 1 will cover cutting-edge academic science and day 2 will focus on translational, clinical, and regulatory science. The conference aims to bridge the gap between basic scientists, clinicians, regulators, and industry. Providing a platform for discussing new findings and facilitating networking among academic and translational researchers, industry professionals, regulators, and funders.
duet multiomics solution evoC
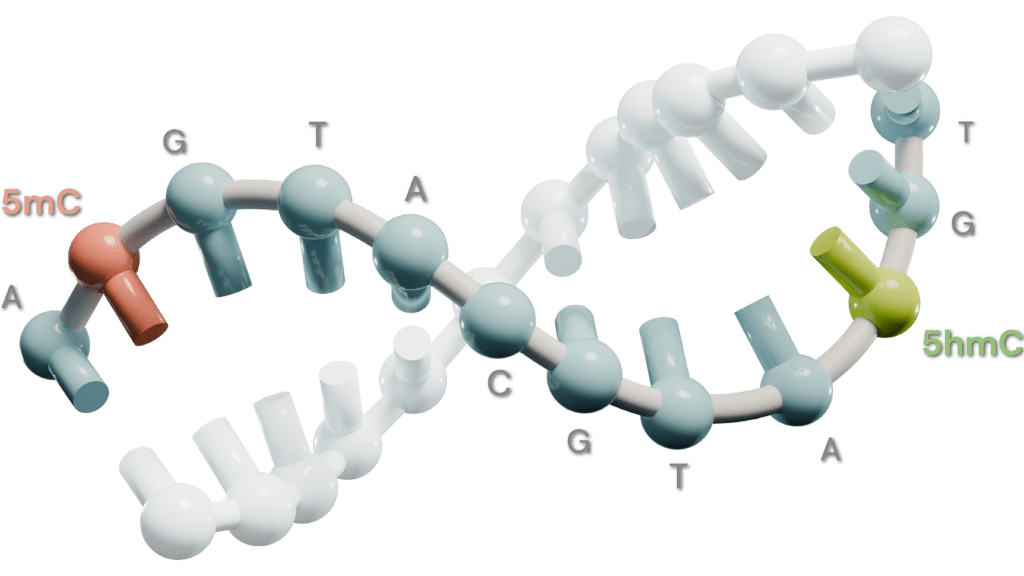
Unlock the power of the 6-base genome with duet evoC data, providing unprecedented insight, including A, C, G, T plus 5mC and 5hmC, in a single read. Learn more