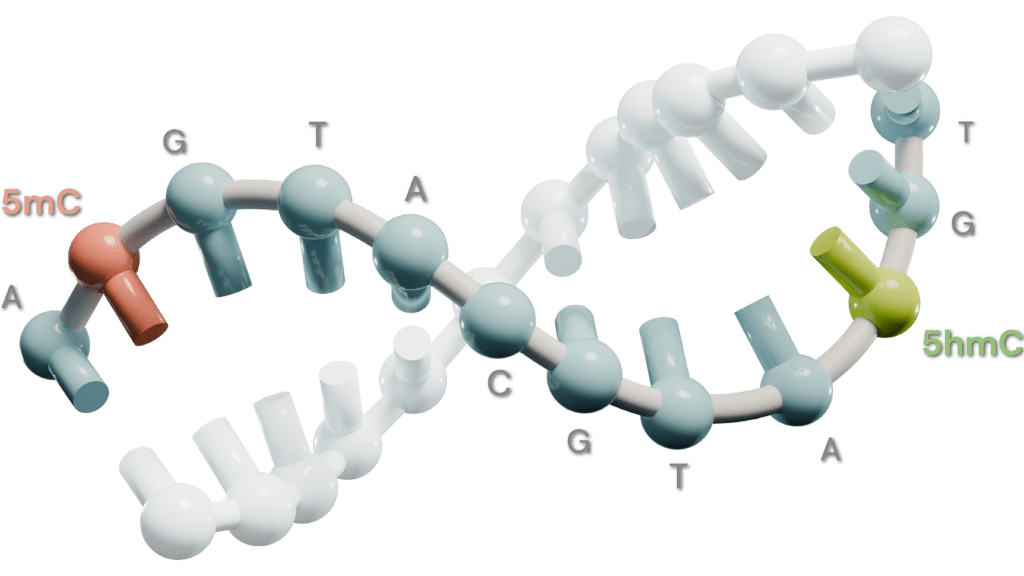
A key challenge in genomics is the generation and integration of data across different modalities. This typically requires several assays to be performed, which is costly and time-consuming, and the subsequent integration of these data across assays is technically challenging. Here, we leverage an assay that enables sequencing the complete genetic sequence and the DNA modifications, 5-methylocytosine (5mC) and 5-hydroxy-methylocytosine (5hmC), from low nanogram amounts of DNA, to provide 6-base genomic data.
Given 5mC and 5hmC play key roles in gene regulation and chromatin organisation, we aimed to explore how these multimodal data could further elucidate key biological processes and yield novel insight. To this end, we trained and evaluated a series of machine-learning models to predict gene expression, chromatin accessibility, and enhancer state from 6-base sequence data.
We use 6-base data from a mouse embryonic stem cell-line, ES-E14TG2A, alongside publicly available polyA RNA-seq, ATAC-seq, TT-seq, and histone modification data of the same cell line as training data and evaluate the performance of the models on held-out test chromosomes. We show that these models can generate highly accurate predictions of gene expression (polyA RNA-seq prediction: R2=0.75, Spearman’s 𝜌;=0.86; TT-seq prediction: R2=0.85, Spearman’s 𝜌; =0.91) and chromatin accessibility (ATAC-seq prediction: R2=0.83, Spearman’s 𝜌;=0.93). Importantly, we found stronger performance of our models for predicting TT-seq over polyA RNA-seq signal, suggesting that 6-base data offers a powerful window into nascent transcriptional activity. As well as being able to predict continuous expression and chromatin accessibility metrics, we show 6-base data predicts enhancer state (Active, Primed, Repressed), defined by histone modifications, with 91% accuracy. In all models the addition of resolved 5hmC signal over undifferentiated 5mc and 5hmC improved predictive performance.
We have shown that the combination of resolved methylation and genomic data combined with machine-learning can generate accurate inference of other data modalities which play key roles in gene regulation. Thus, there is a compounding effect whereby 6-base genomic assays not only yield direct data, but also the foundations for multiple other inferred modalities. Looking ahead, these approaches can enable novel insights into core biological processes and accelerate the speed of iteration for experimental projects, where one can yield multifaceted insights and even conduct pilot experiments in silico using predictive models.