- Robert Crawford
- Ben Krajacich
- Ermira Lleshi
- Jack Monahan
- Fabio Puddu
- Jamie Scotcher
- Aurel Negrea
- Lisabet Andreasen
- Jens Fullgrabe
- David Morley
- Donna McDade Walker
- Semyon Kruglyak
- Páidí Creed
The combinatorial power of genetics and epigenetics is vital to understanding biology in healthy and cancerous states. Utilising a recent novel approach (duet multiomics solution evoC), we enable the simultaneous identification of modified cytosine (both 5mC and 5hmC) and the canonical bases A, C, T & G in an enzymatic single-workflow solution. Generating this information across the whole genome provides a much-improved understanding of genetic and epigenetic changes, but can require a large amount of sequencing, particularly when exploring at depth. Enrichment protocols such as target enrichment enable deep interrogation of target areas whilst maintaining cost effectiveness.
Target enrichment panels enable the selective isolation of sequences of interest through target hybridisation. Here we sequenced low input cell-free DNA (cfDNA) duet evoC libraries targeted with a commercially available panel (Twist Human Methylome) on Element Aviti using Direct linear library loading with Cloudbreak FreeStyle Sequencing. Output data resulted in highly accurate genetic and epigenetic information for regions of interest with reduced sequencing requirements. Samples used were cfDNA from healthy patient samples and patients with either Stage I or IV Colorectal Cancer (CRC). Widespread differences in methylation and hydroxymethylation are clearly observed in late-stage cancer vs early-stage or healthy controls demonstrating utility of the approach for applications such as liquid biopsy.
Refining genetic and epigenetic investigations to a narrower region of interest can improve detection of low prevalence disease-associated biomarkers at significant sequencing depth and lower sequencing costs.
- Strand synthesis: creates a single molecule with a direct copy of the original information tethered together with a hairpin. The copy strand is without cytosine modifications initially, but importantly, utilises a high fidelity methyltransferase to copy over only 5mC from the original to the copy strand.
- Sequencing paired-end read: generates sequence information after protection of cytosine modifications followed by deamination of all remaining cytosines (read as thymine in NGS).
- Read resolution: aligns original and copy strands to correctly call all 4 canonical bases along with 5mC and 5hmC.
- Aligned (4 base) reads with 5mC & 5hmC are tagged (6 base information)
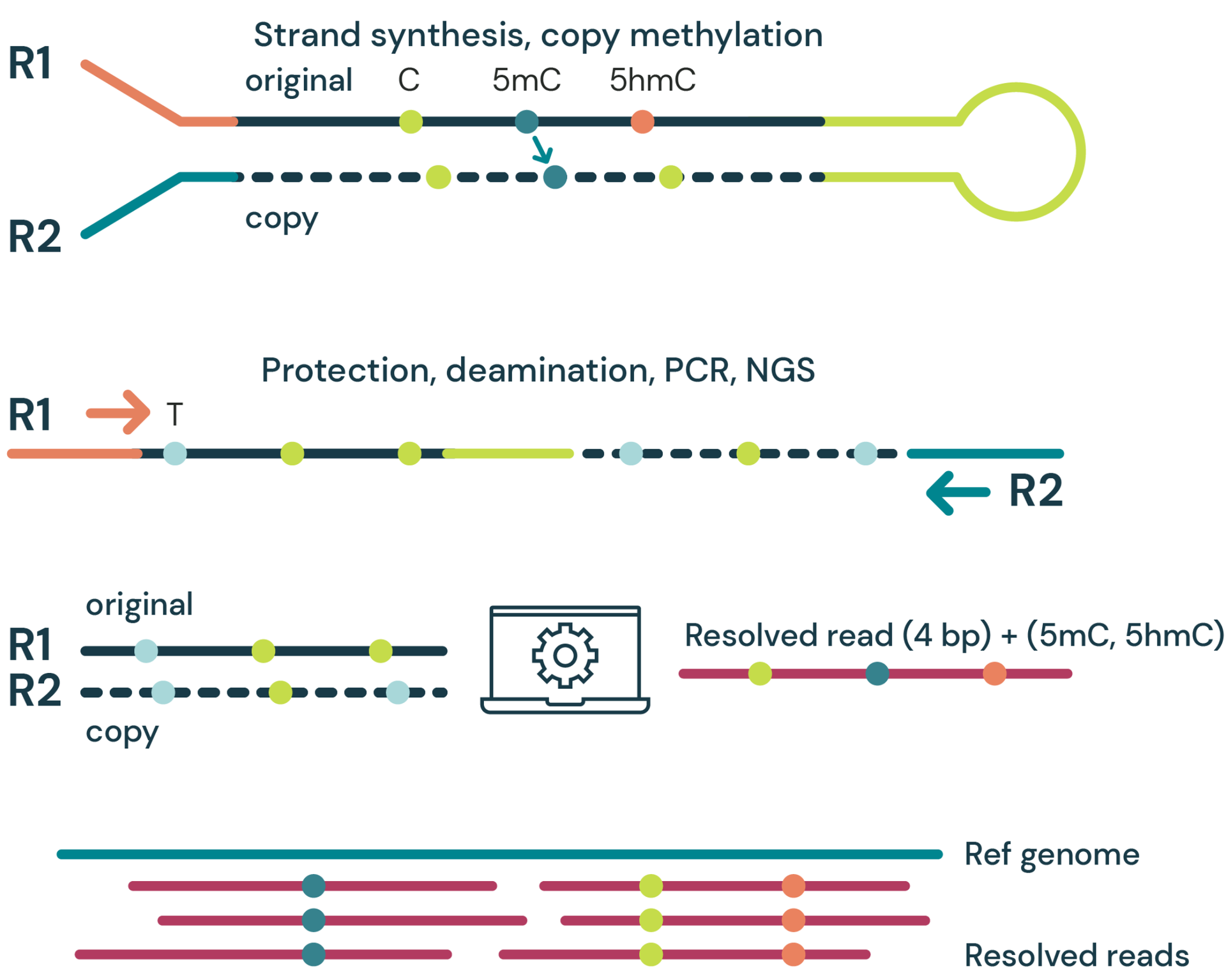
duet multiomics solution evoC is a 6-base calling technology that reads all four canonical bases plus 5mC and 5hmC.
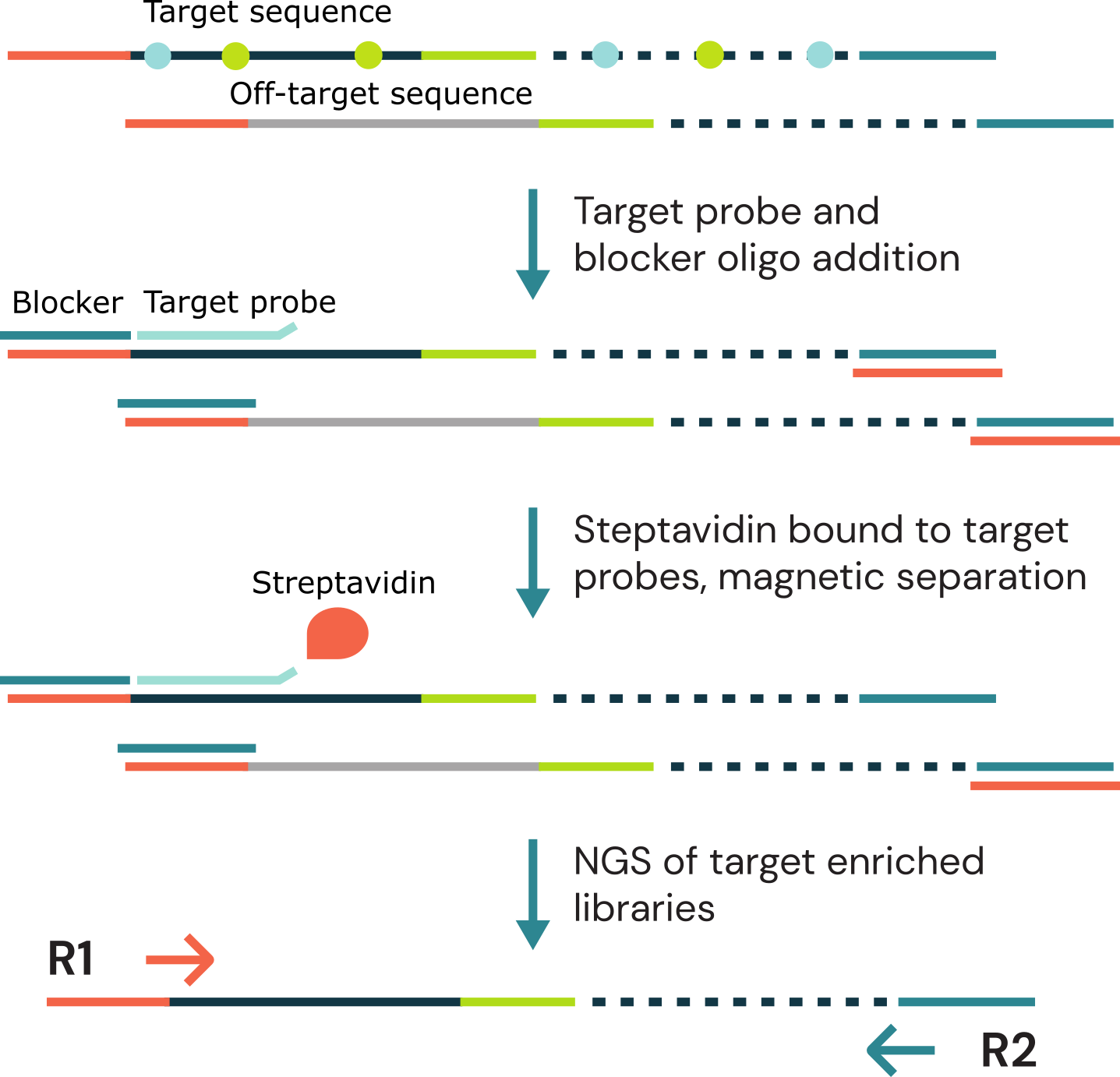
Workflow schematic for duet evoC target enrichment
Whole genome duet evoC libraries are pooled, then hybridised to target probes and blocking oligos. Captured DNA/probe complexes are then bound to streptavidin beads and amplified by PCR, followed by a final bead clean-up.
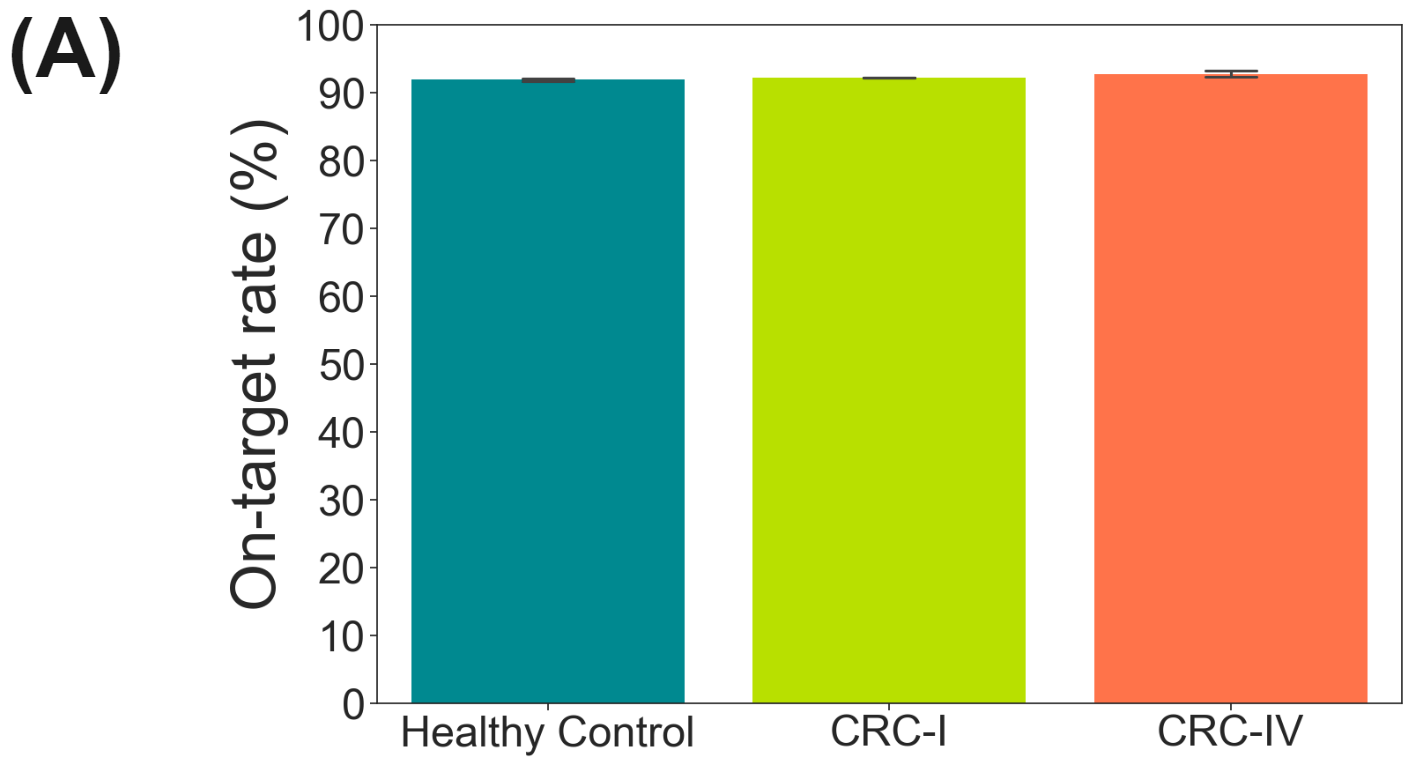
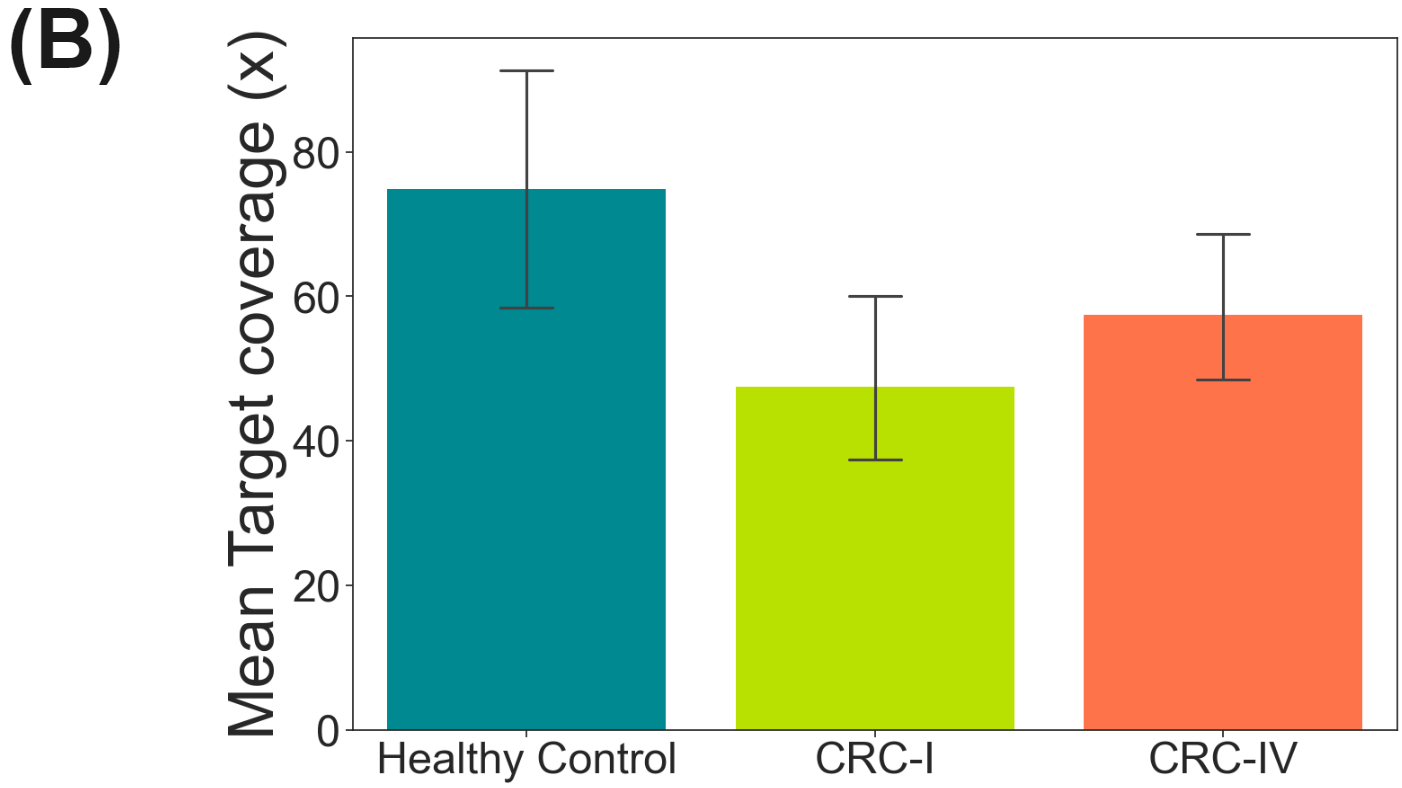
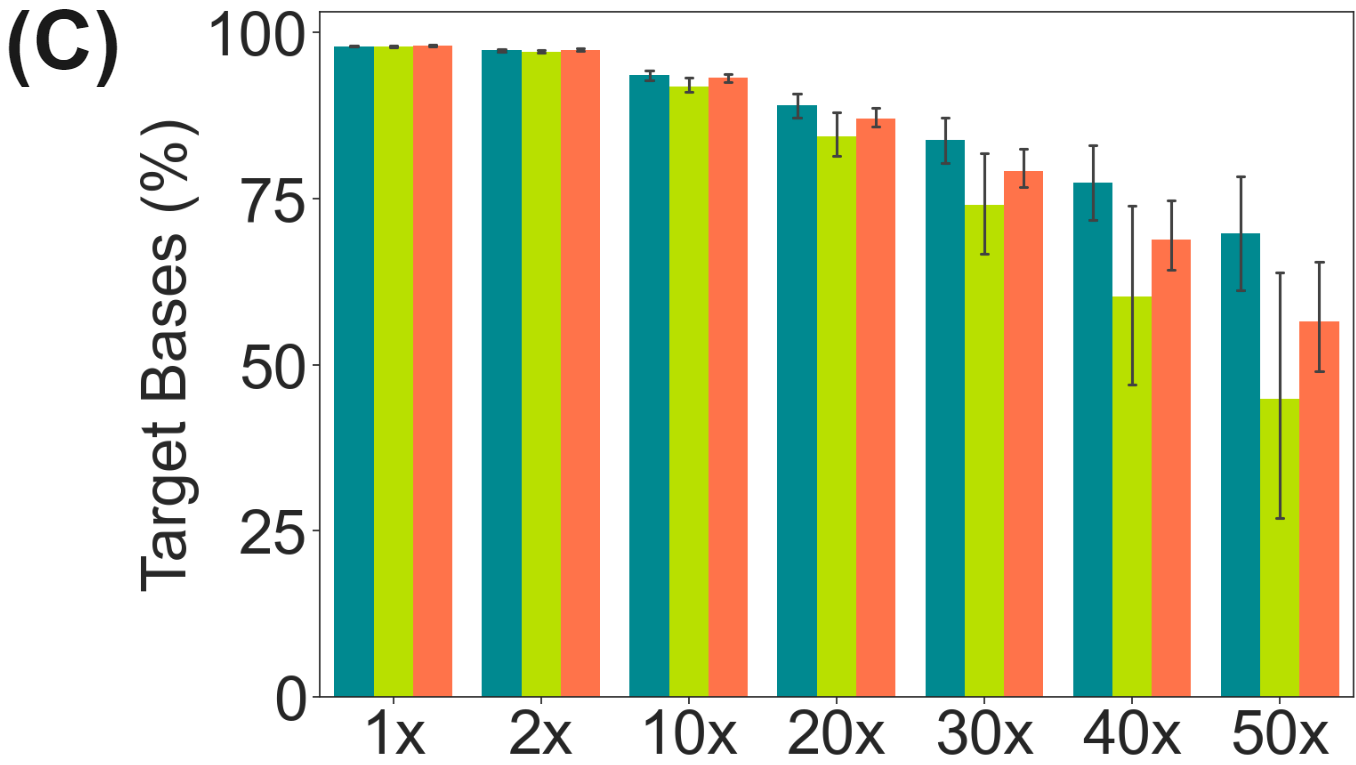
duet evoC targeted with Twist Human Methylome panel.
Here, a pool of 10ng cfDNA duet evoC libraries (Healthy Control, CRC-I & CRC-IV) were targeted using the Twist Human Methylome panel and sequenced on Element Aviti (~2B reads total, @ 94.7-95.3% Q30).
- On-target rate was ~92% for all samples.
- Mean target coverage was ~75X, 48X & 57X for Healthy Control, CRC-I and CRC-IV respectively.
- 74-84% of methylome targets are covered at 30X or higher.
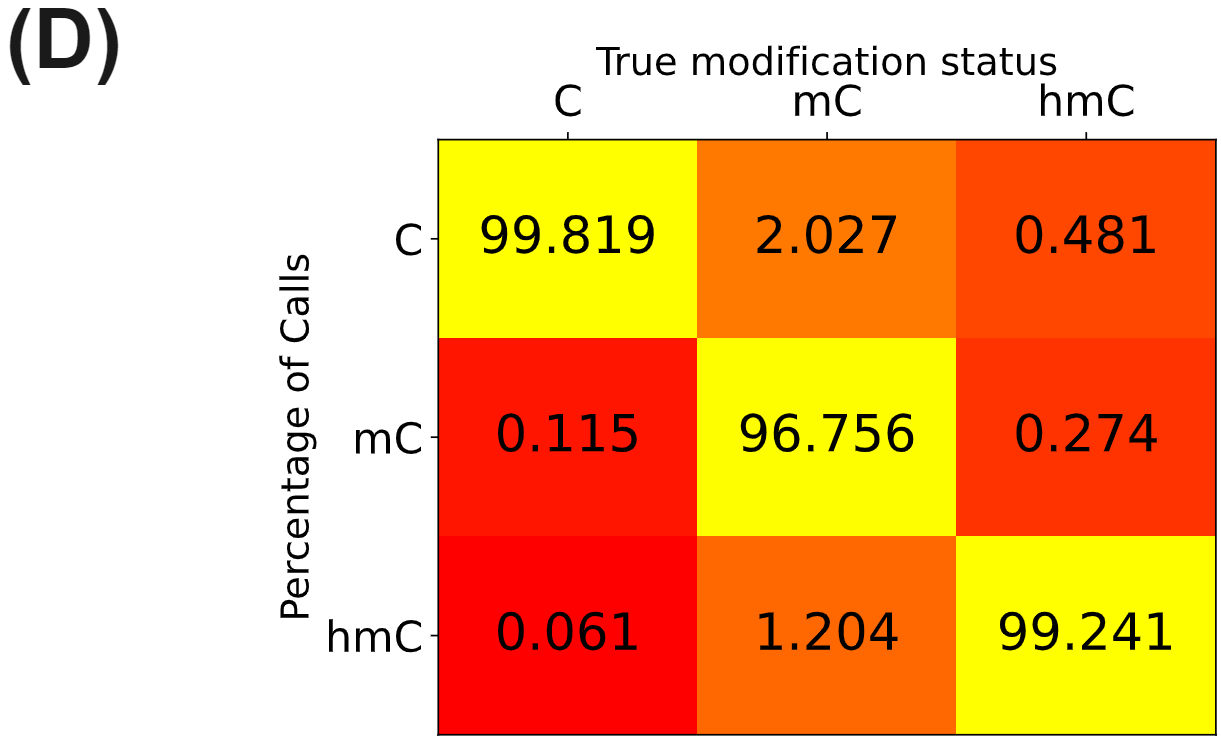
- Call-rate matrix containing the rate at which duet evoC with Element Aviti calls unmodC, 5mC and 5hmC from control sequences with known modification status. Samples were prepared with strand-reversing, IDT xGen UDI-UMI adapters. UMI-aware deduplication and targeted performance metrics analyses were performed with picard. All figures use data generated from cfDNA extracted from plasma of patients with colorectal cancer or healthy volunteers.
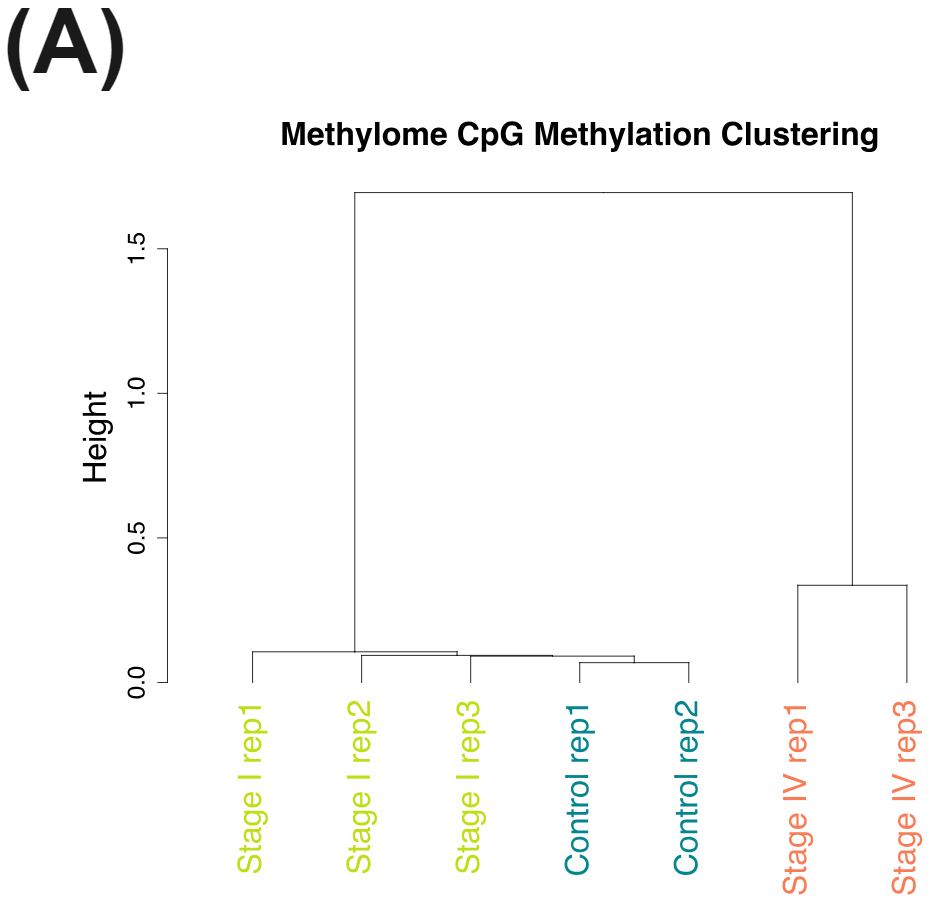
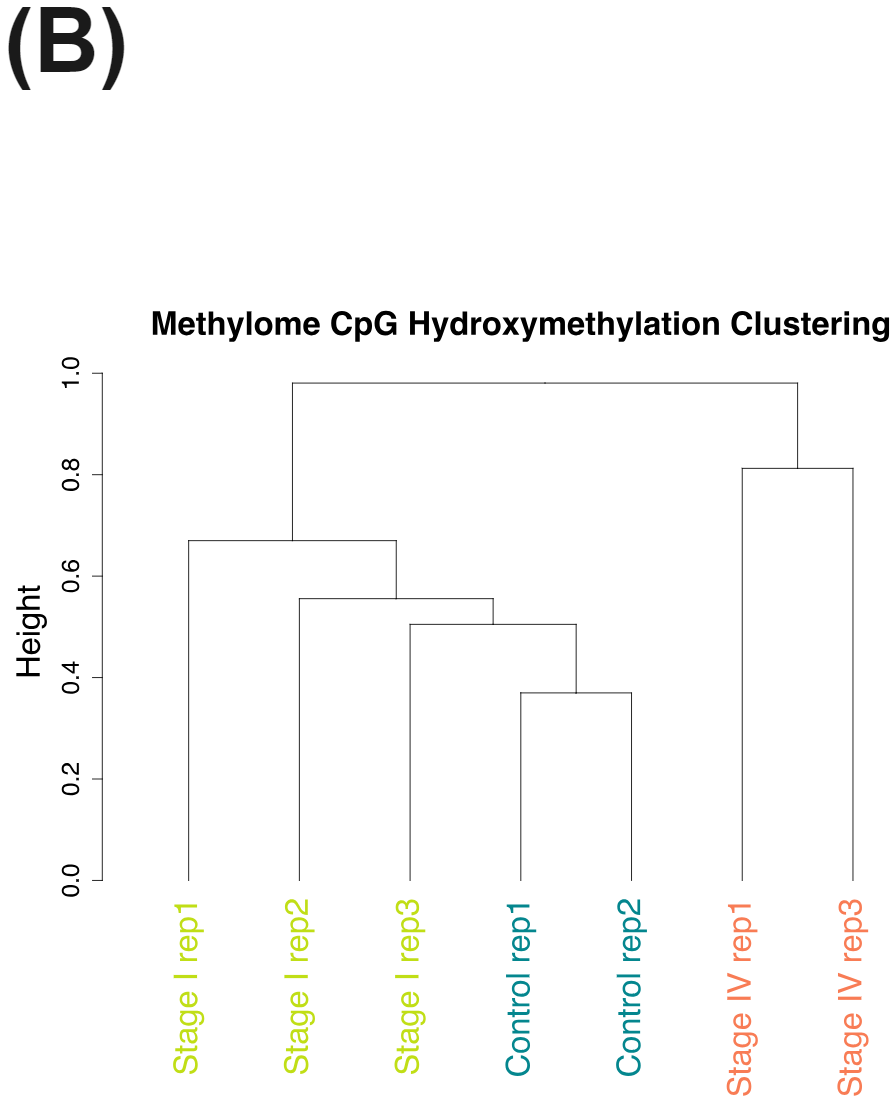
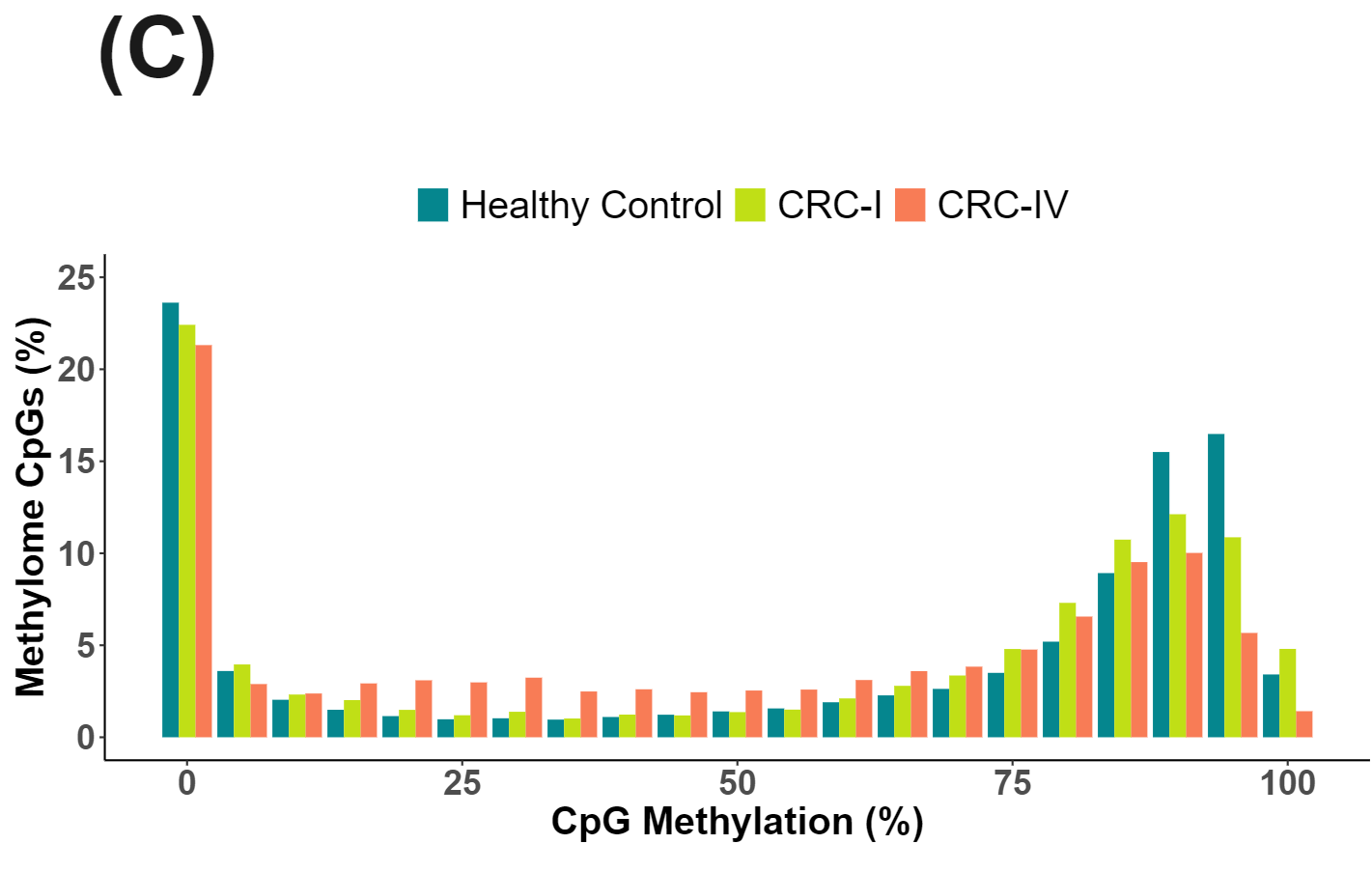
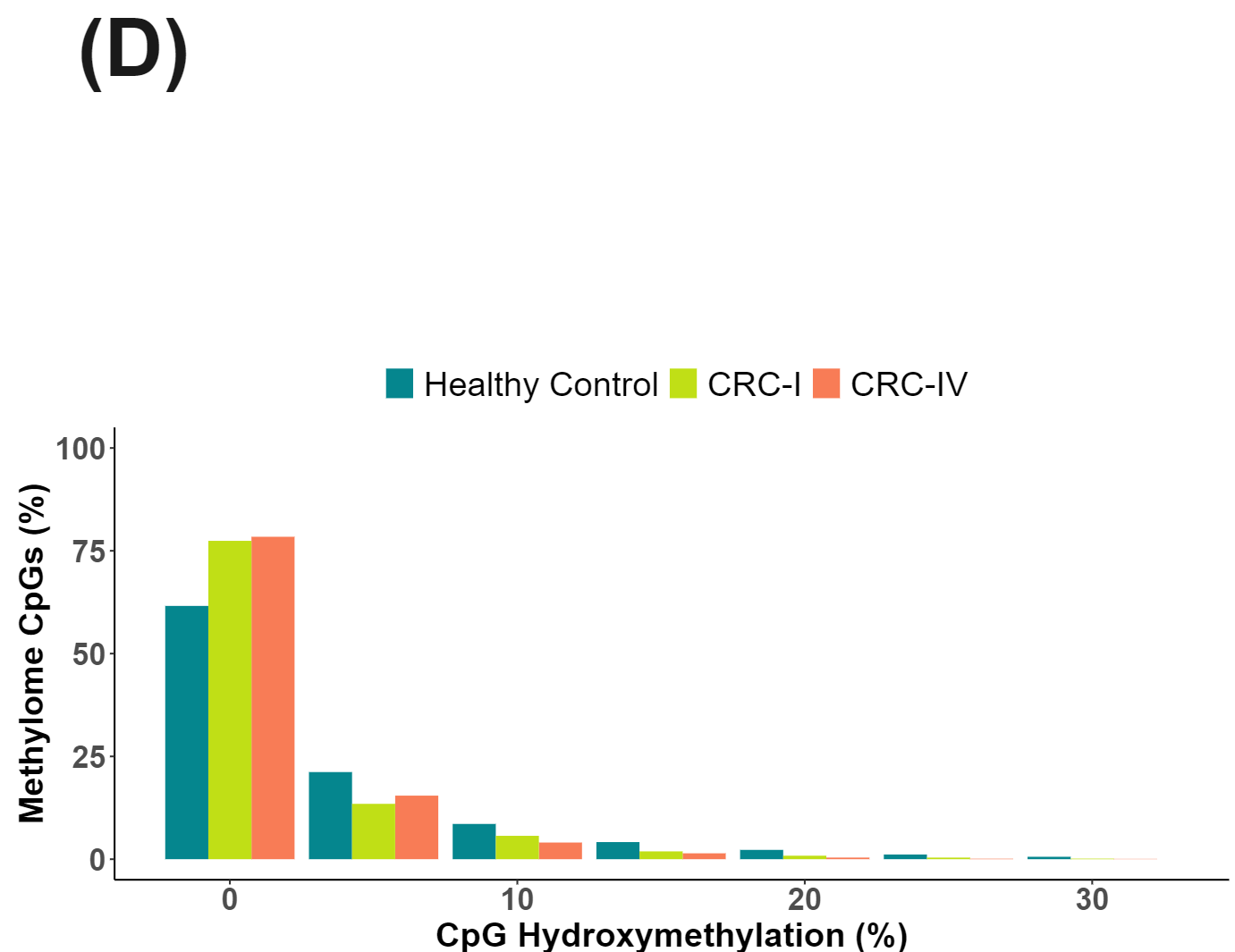
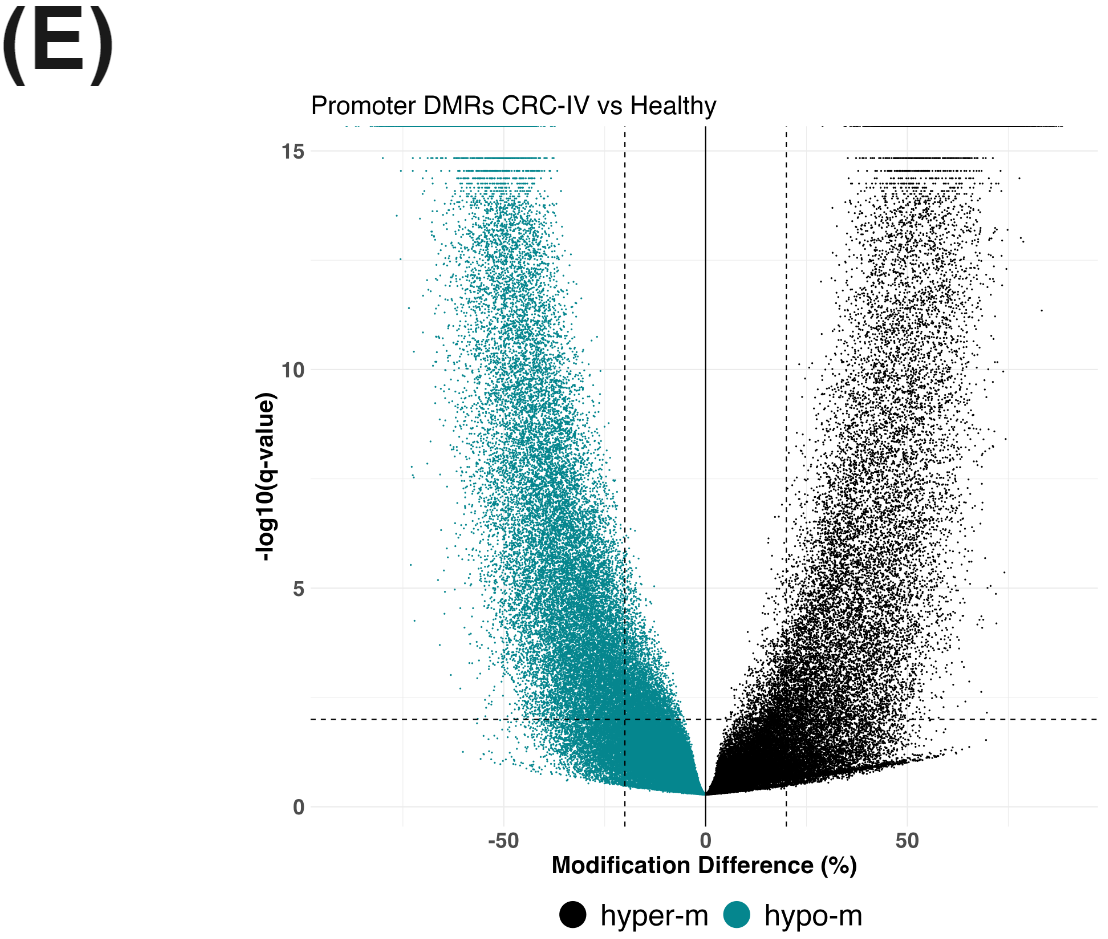
cfDNA from patients with colorectal cancer and healthy volunteers have distinct patterns of cytosine methylation and hydroxymethylation at CpGs.
cfDNA is shed into the bloodstream during programmed or necrotic cell death. Most cfDNA will originate from healthy cells with a very small fraction being circulating tumour DNA (ctDNA).
Nevertheless the cfDNA from the late stage CRC patients have very distinct methylomes (A) and hydroxymethylomes (B) that cluster seperately to cfDNA samples obtained from early stage CRC patients and healthy volunteers.
There is significant reduction in CpG methylation in the Methylome Panel with CRC oncognesis and disease progression (C).
Hydroxymethylation is less abundant than methylation in the methylome panel but nonetheless shows an increase in overall hypo-hydroxymethylation for the disease states (D).
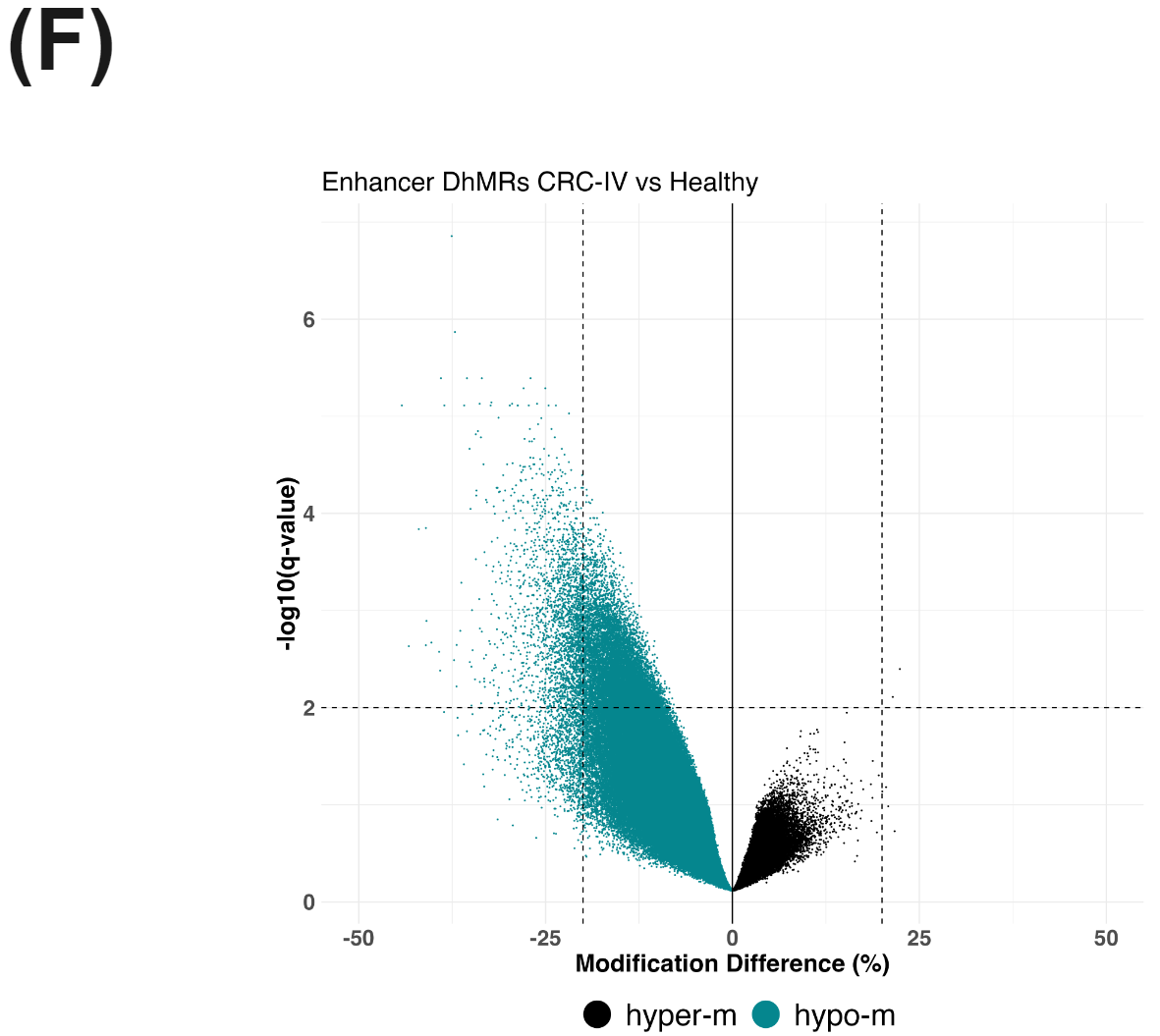
There is a substantial number of differentially methylated CpGs detected in promoter regions (methylKit using beta-binomial model) comparing CRC-IV to healthy controls (E), likely reflecting a mix of hypo- and hyper-methylation accompanying oncogenesis. In parallel, we see evidence for hypo-hydroxymethylation at enhancer elements (F).
All figures use data generated from cfDNA extracted from plasma of patients with colorectal cancer or healthy volunteers
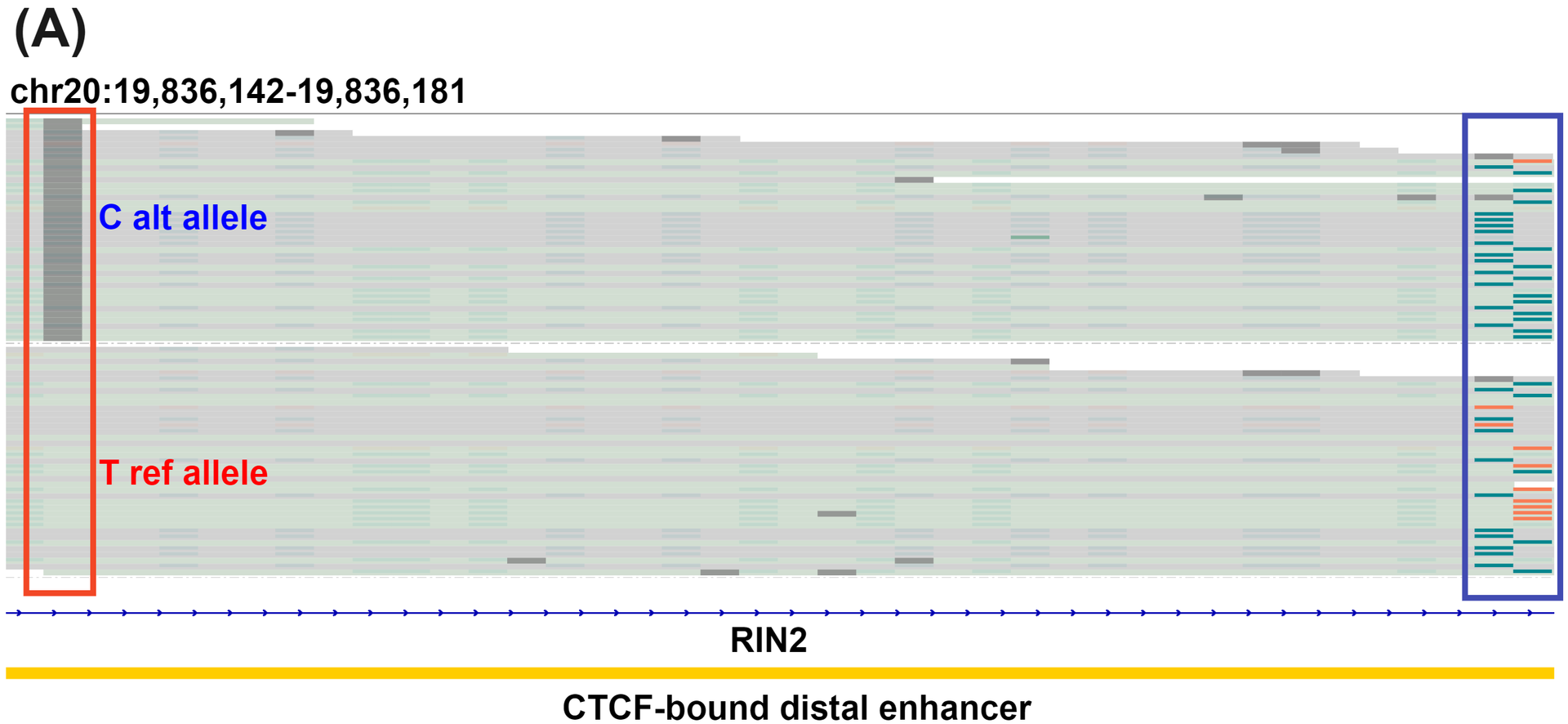
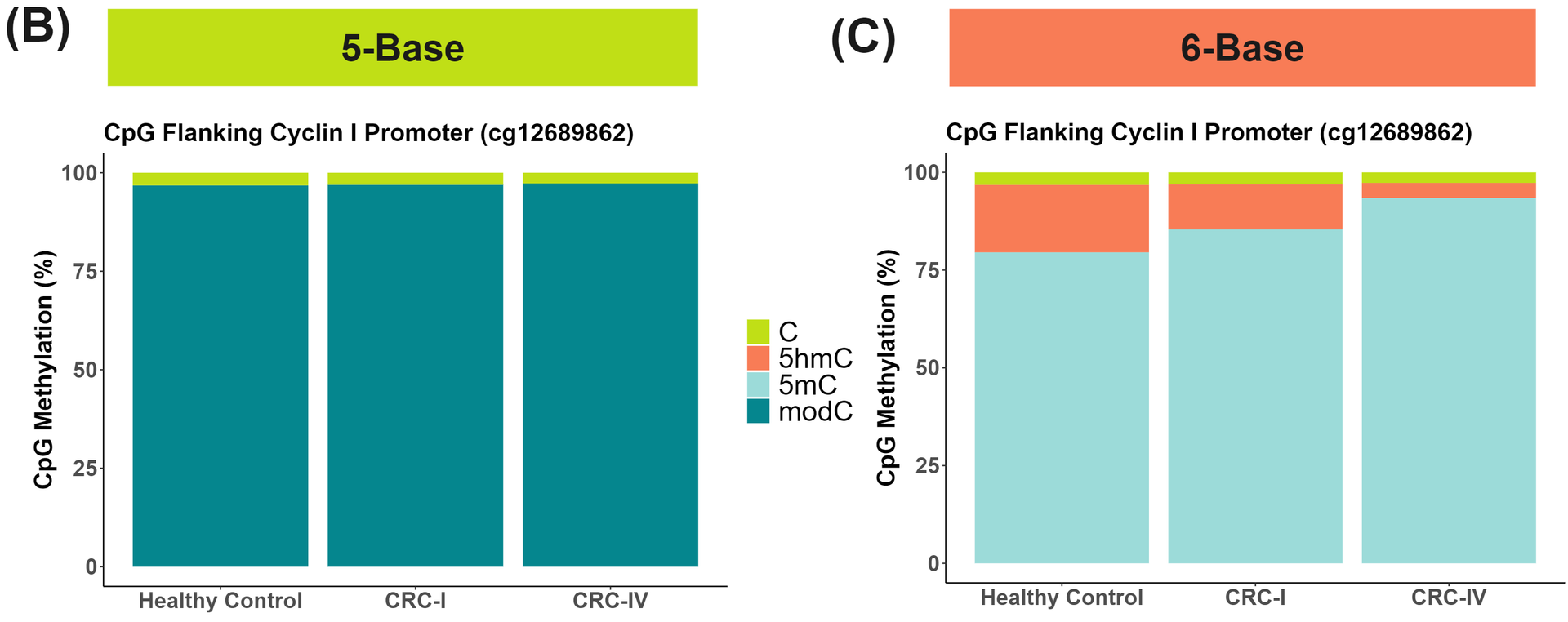
Target enriched duet evoC detects Variant Associated Methylation (VAM) and identifies transitions in cytosine methylation occurring in CRC progression.
duet evoC detects allele specific hydroxymethylation (ASM), a form of VAM, within an intronic CTCF-bound enhancer in RIN2, a member of the Ras association domain family of tumour suppressors that frequently undergo epigenetic inactivation in CRC (A). The C alt allele is associated with decreased 5hmC (orange) and increased 5mC (teal). Resolving methylation (B) in a 5-Base space masks distinct 5mC and 5hmC trajectories in the promoter flanking region of Cyclin-I, a potential CRC neoantigen. These distinct modalities are revealed when we resolve (C) in a 6-Base space allowing detection of otherwise cryptic 5mC and 5hmC transitions.




