Authors: A. Tivey, A. Clipson, M. Halford, D. A. Onuselogu, P. Harker, M. Hossain, S. M. Hill, S. Makeev, D. Rothwell, C.Dive, R. Lee, F. Mouliere
Patient outcomes in advanced melanoma have been transformed by immune checkpoint inhibitors (ICIs). Patients who develop resistance have limited treatment options; there is a need for biomarkers to identify these patients and refine treatment decisions.
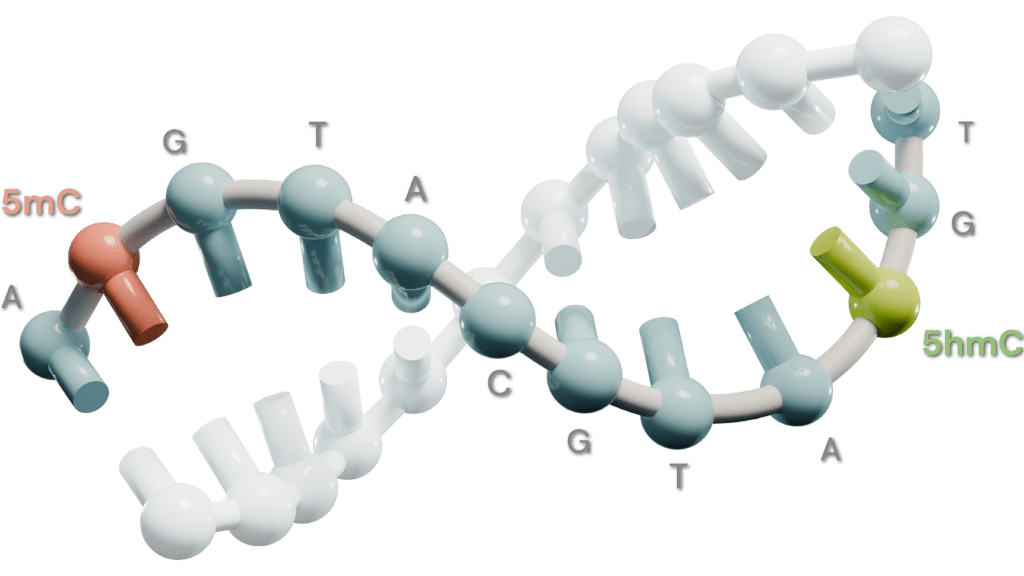
A multi-omic cell-free DNA (cfDNA) based ‘liquid biopsy’ could offer insight into tumour and immune changes at baseline and throughout therapy. Genomic analyses could inform on tumour mutational signal and neo-antigen load. Methylation analyses can be used to deconvolute the cfDNA cell-of-origin which may inform on immune cell turnover and normal tissue toxicity. Multi-omic profiling could enhance liquid biopsy but can involve multiplication of experiments and workflows reducing its potential to be clinically implemented.
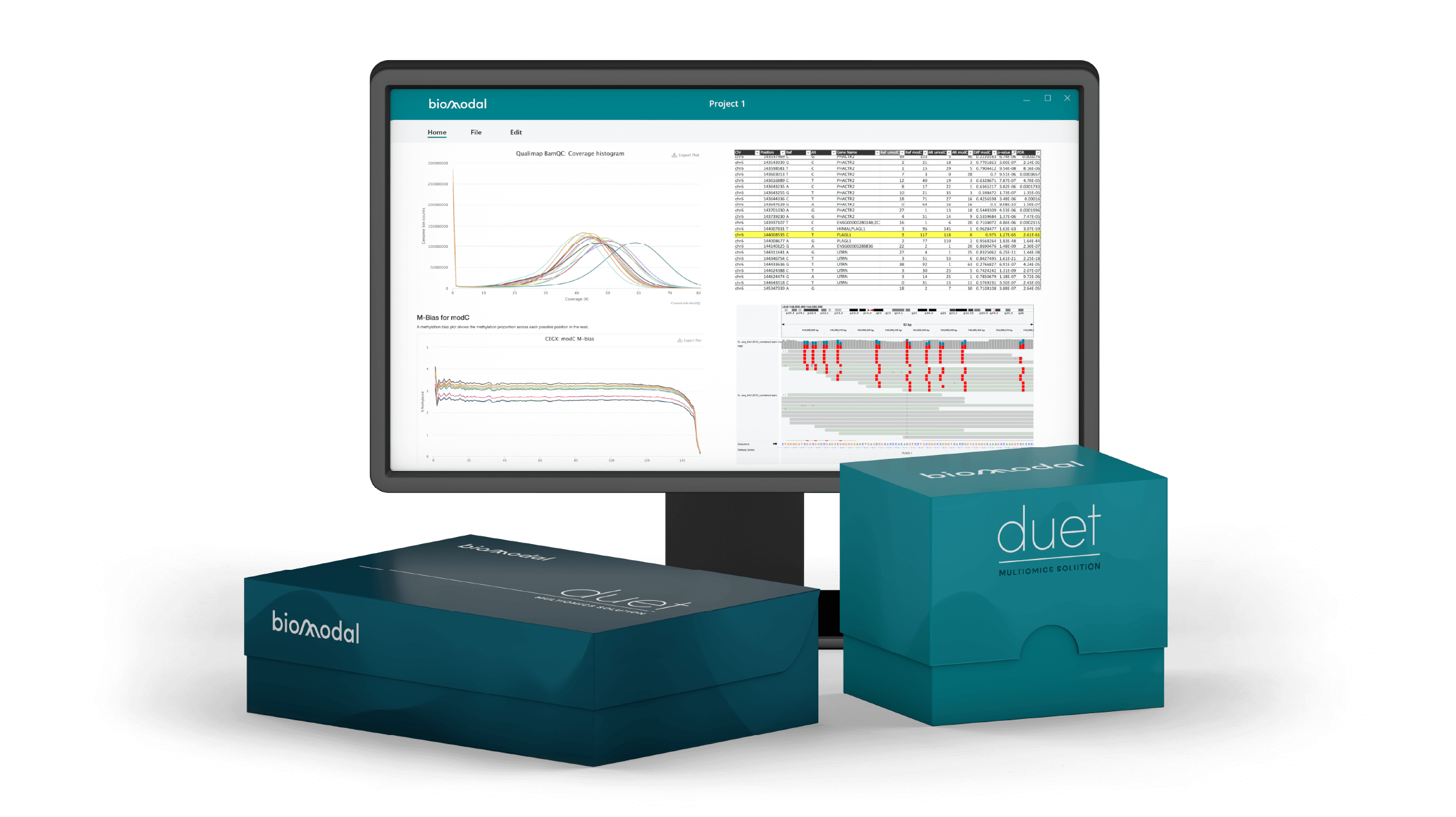
We are evaluating the duet evoC technology which notably provides cfDNA genomic, methylation and hydroxymethylation information from a single workflow. We are comparing this 6-base genome sequencing with targeted sequencing and capture-based methylation enrichment (T7-MBD-seq). Initial analysis of cfDNA samples from patients with advanced cancer has shown concordance of tumour fraction and methylation profiling when assessed by the different methods. At low coverage (~3x) WGS, 88% mutations with variant allele frequency of ≥20% by targeted sequencing were correctly identified by duet evoC. Both de novo identified differentially methylated regions and regions from an external reference atlas were able to distinguish samples from patients with melanoma from other samples using this technology.
Future objectives include comparing duet evoC with T7-MBD-seq for deconvoluting cell-of-origin from in-vitro and in-silico mixture sets of reference material, and comparing the methylation and hydroxymethylation profiles in longitudinal samples from patients with melanoma treated with ICIs.
In conclusion, we are assessing the potential of a multi-omic assay that can capture both genomic and epigenomic features of tumour- and immune-derived cfDNA. This will enable us to characterise comprehensively the cfDNA phenotypes associated with ICI response and resistance in patients with melanoma.