Formalin-fixed, paraffin-embedded (FFPE) samples are a well known and widely used sample type in research and clinical settings, offering long-term storage for applications such as oncology and genomics. Such samples have great potential to add value to studies incorporating both genomic and epigenomic information such as the cytosine modifications 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), providing far greater insights than WGS or immunostaining alone. For example, epigenetic insights into gene activation and silencing pathways enable a better picture of gene regulation in cancer. However, formalin fixation can cause DNA damage, such as deamination, fragmentation and crosslinking, reducing data quality in next-generation sequencing (NGS).
Here we apply 6-base sequencing using duet evoC sequenced on the NovaSeqX to both formalin-damaged standards and clinical fresh frozen (FF) and FFPE samples from colorectal and lung cancers. 6-base sequencing enables the simultaneous detection of modified cytosines (5mC, 5hmC) and canonical bases (A, C, G, T) in a single workflow, minimising information loss and preserving the ability to detect common somatic C-to-T transitions. This study investigates the impact on 6-base data quality of formalin damage in such samples and compares to traditional methods that provide a conflated modC readout.
Fragmented DNA is ligated to a hairpin adapter at both ends. Hairpin complex is split into two strands and a complementary copy strand is synthesized, accurately capturing the genetic sequence prior to conversion. 5mC is copied from the original strand to the copy strand whereas 5hmC does not get copied.
5mC and 5hmC on both strands are protected, unmodified cytosines are deaminated to uracil.
After PCR and paired end sequencing complementary reads are pairwise aligned. Bases from both reads are resolved to identify the original base identity whilst the CpG context across strands is used to distinguish 5mC from 5hmC to yield 6-base data. Sequencing or PCR errors will result in implausible pairs that are identified, labelled N, and filtered.
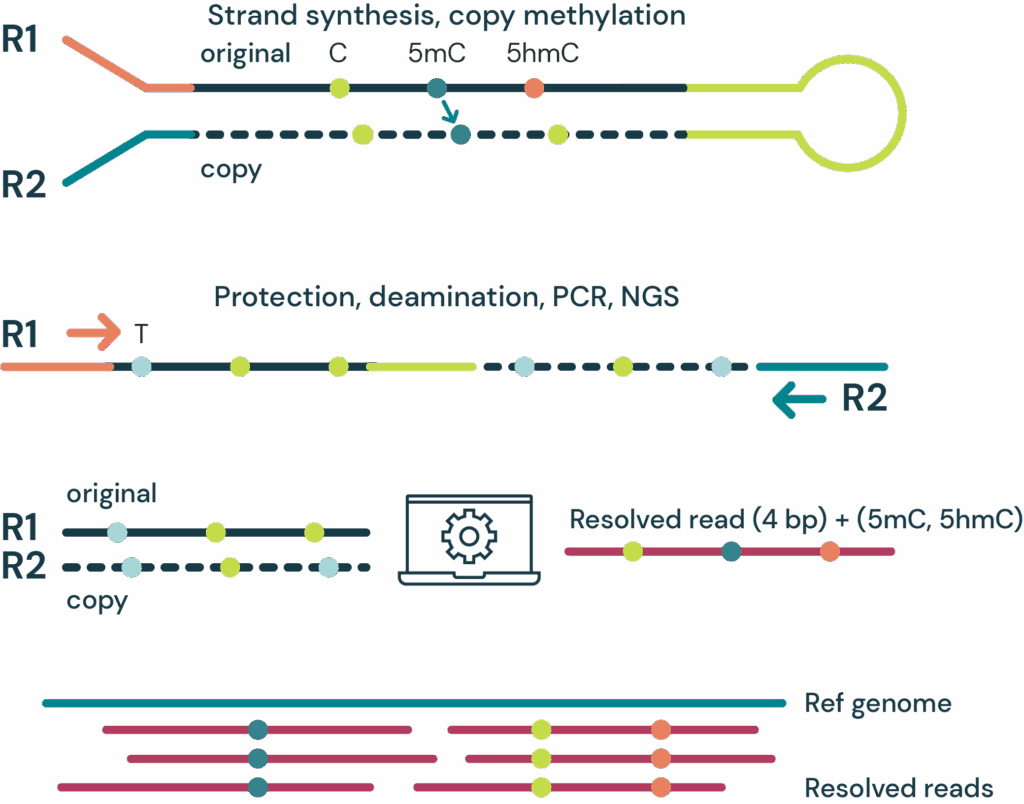
duet evoC is a 6-base sequencing technology that reads all four canonical bases plus 5mC and 5hmC.
duet multomics solution evoC and EM-seq v2 applied to formalin-damaged DNA.
Libraries were generated in duplicate with 80ng of Horizon Quantitative Multiplex Reference Standard (QMRS) DNA using either untreated or formalin-compromised DNA (fcDNA) with mild, moderate and severe damage. QMRS contain clinically relevant mutations, enabling assessment of variant detection alongside epigenetic profiling. In addition, four libraries were generated from matched FFPE and fresh frozen (FF) cancer tissue samples.

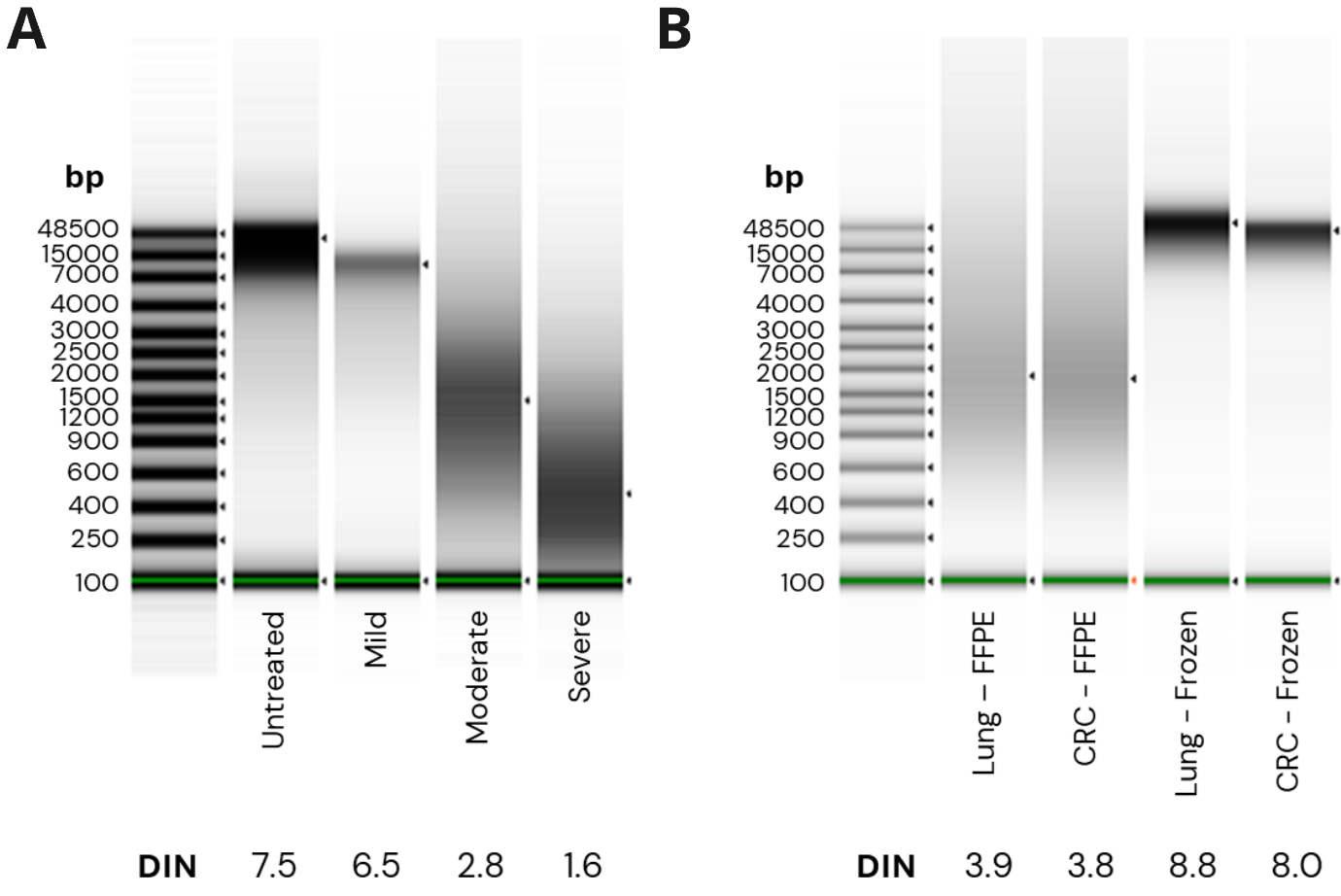
- A. fcDNA quality was assessed using the ScreenTape assay. Mild, moderate and severe damage corresponded to DNA Integrity Number (DIN) ranges of > 5.1, 2.5 – 5.1 and < 2.5 respectively.
- FFPE cancer samples show DIN values of 3.9 and 3.8 and FF samples show DIN scores ≥ 8.
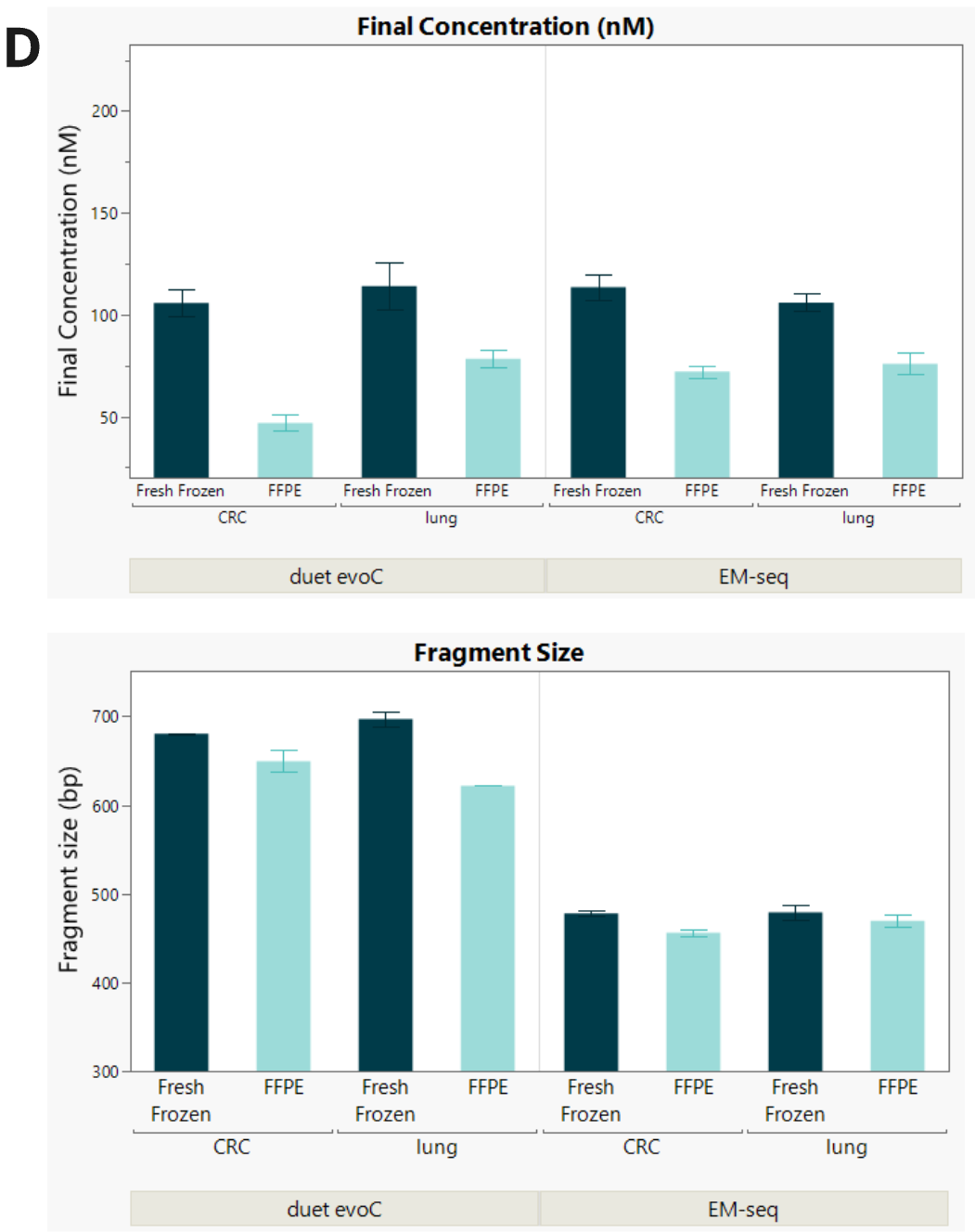
- Additional PCR cycles were required to maintain yields for moderate and severe FFPE processed with duet evoC (+1) and EM-seq v2 (+2) , with yields comparable between technologies. The additional PCR cycle doubled yield for mild fcDNA compared to untreated control. Increasing formalin damage reduced library insert size for both technologies, as expected.
- FFPE libraries have lower yield and insert size compared to matched FF samples for both duet evoC and EM seq v2.
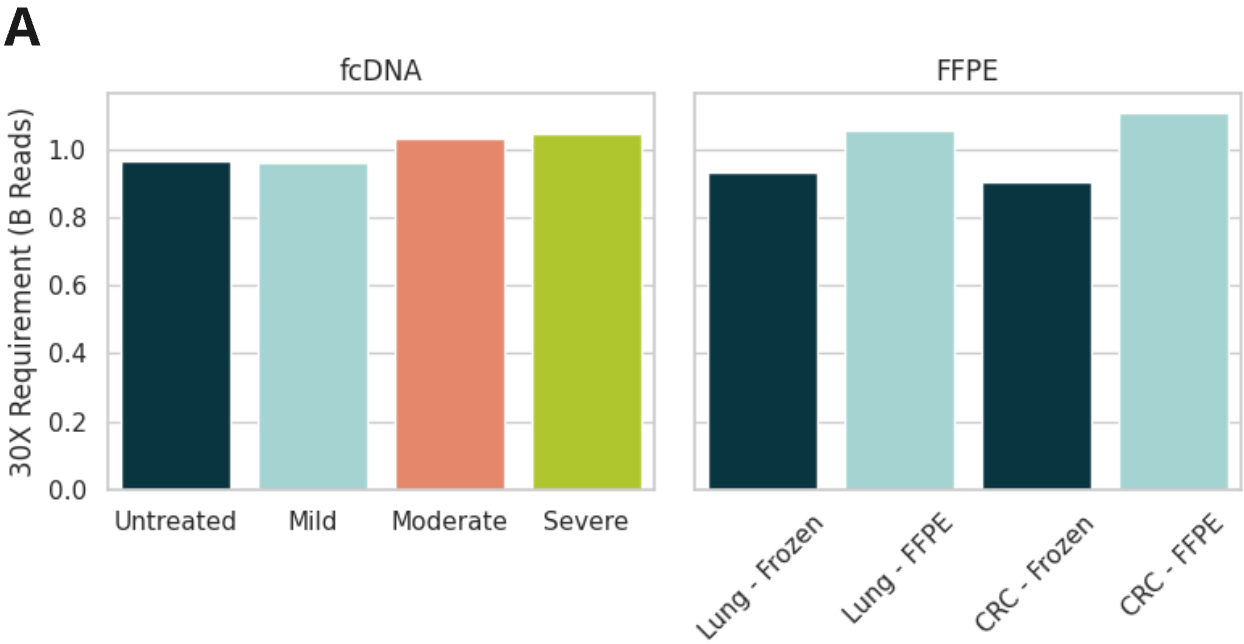
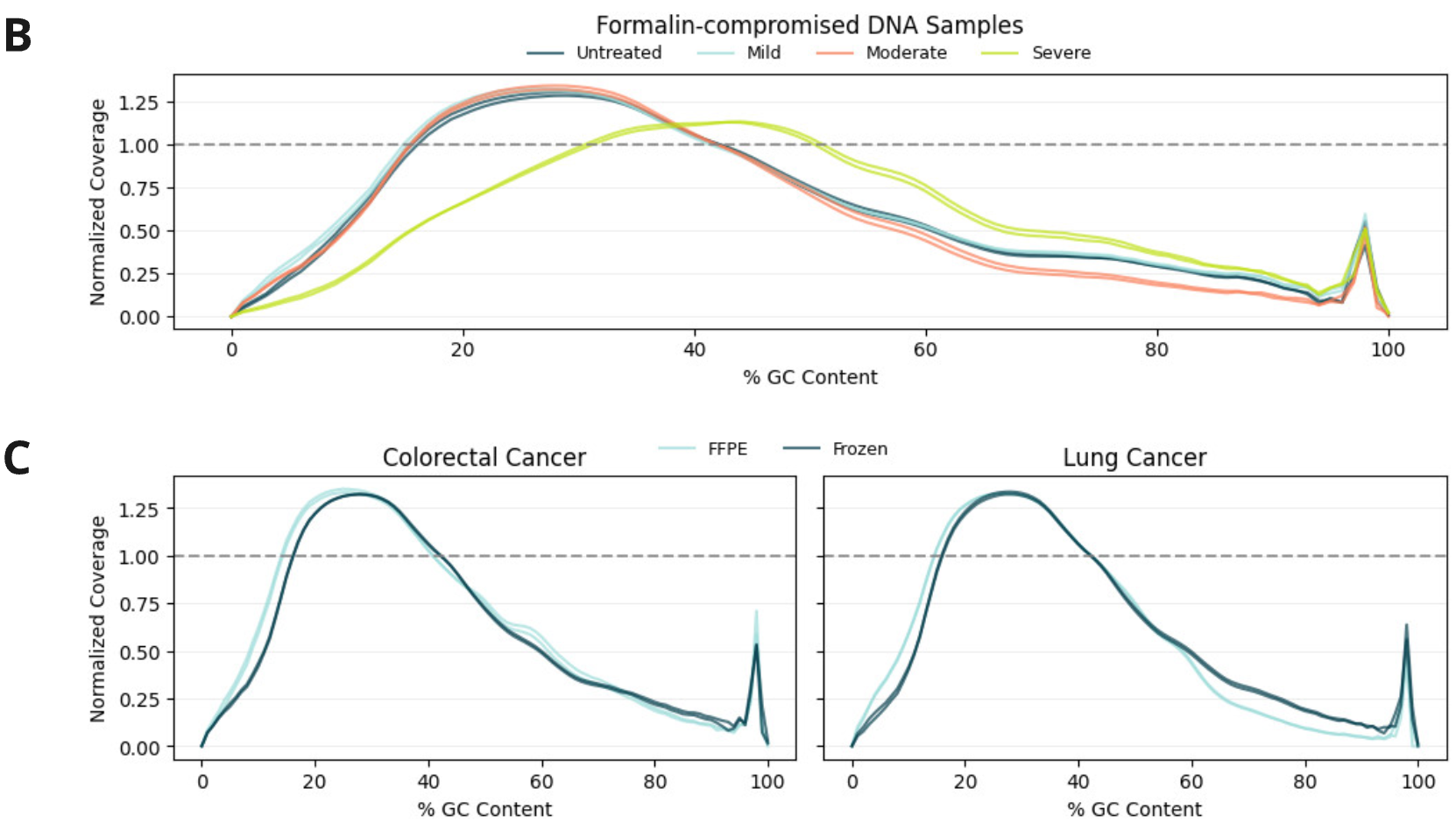
Sequencing performance of duet evoC using fcDNA and FFPE/Fresh Frozen samples after sequencing on Illumina NovaSeq X 25B PE150 kit to ~30x coverage and processing using biomodal’s duet software
- Due to the decreased insert size formalin damage results in a marginal increase in read requirement to achieve 30x depth. An increase of 6.7% reads is required between moderate damage fcDNA and untreated, while an increase of 22.5% and 13.7% reads is required between FFPE and frozen samples in CRC and Lung Cancer, respectively.
- The quality of 6-base data, judged by evaluating GC bias, is robust to and unaffected by mild or moderate formalin damage with an increase in bias only seen in severely damaged samples.
- Similarly the performance of clinical FFPE samples is comparable to FF, indicating that 6-base data between these sample types is equivalent and can be integrated in downstream analysis.
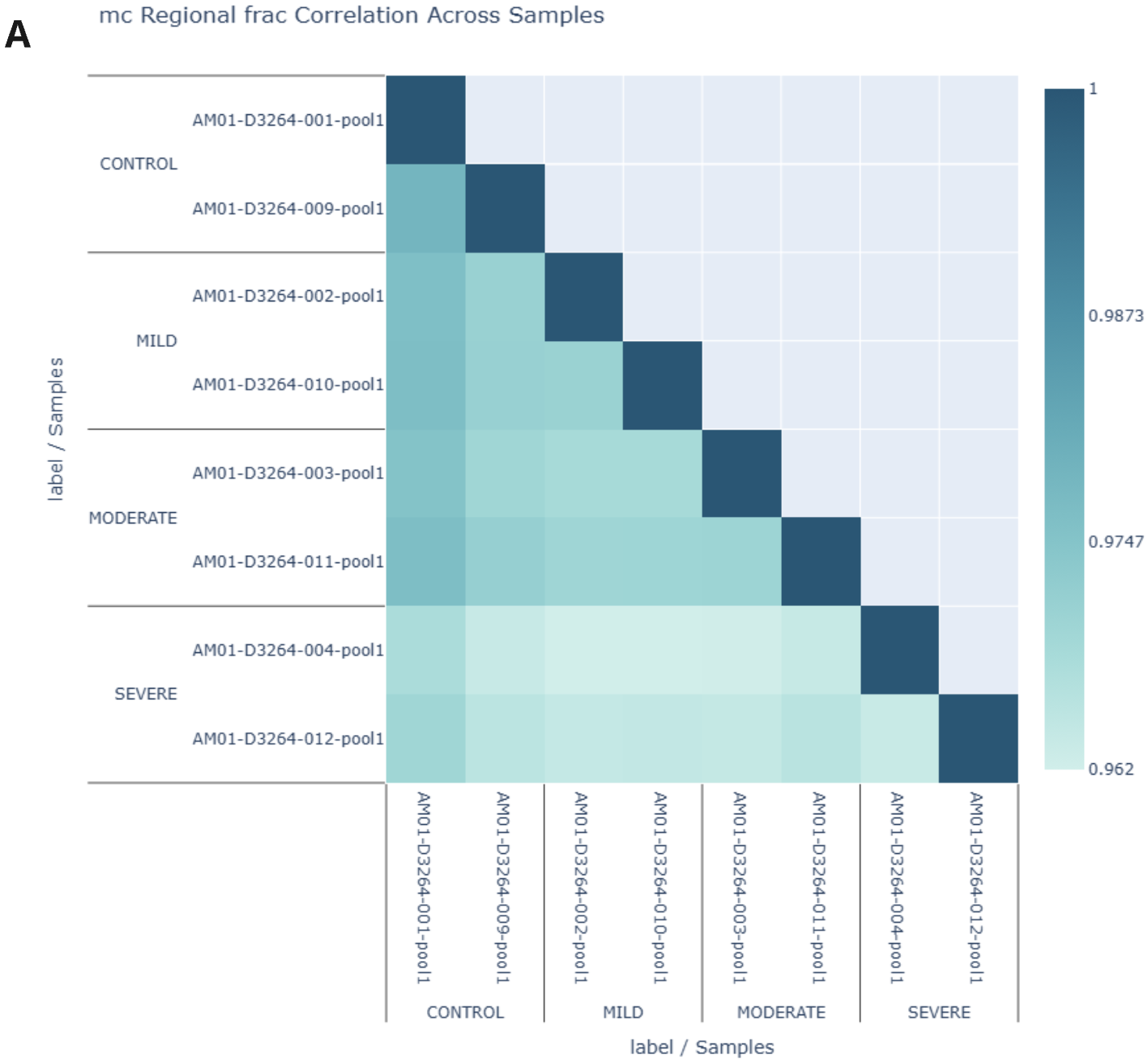
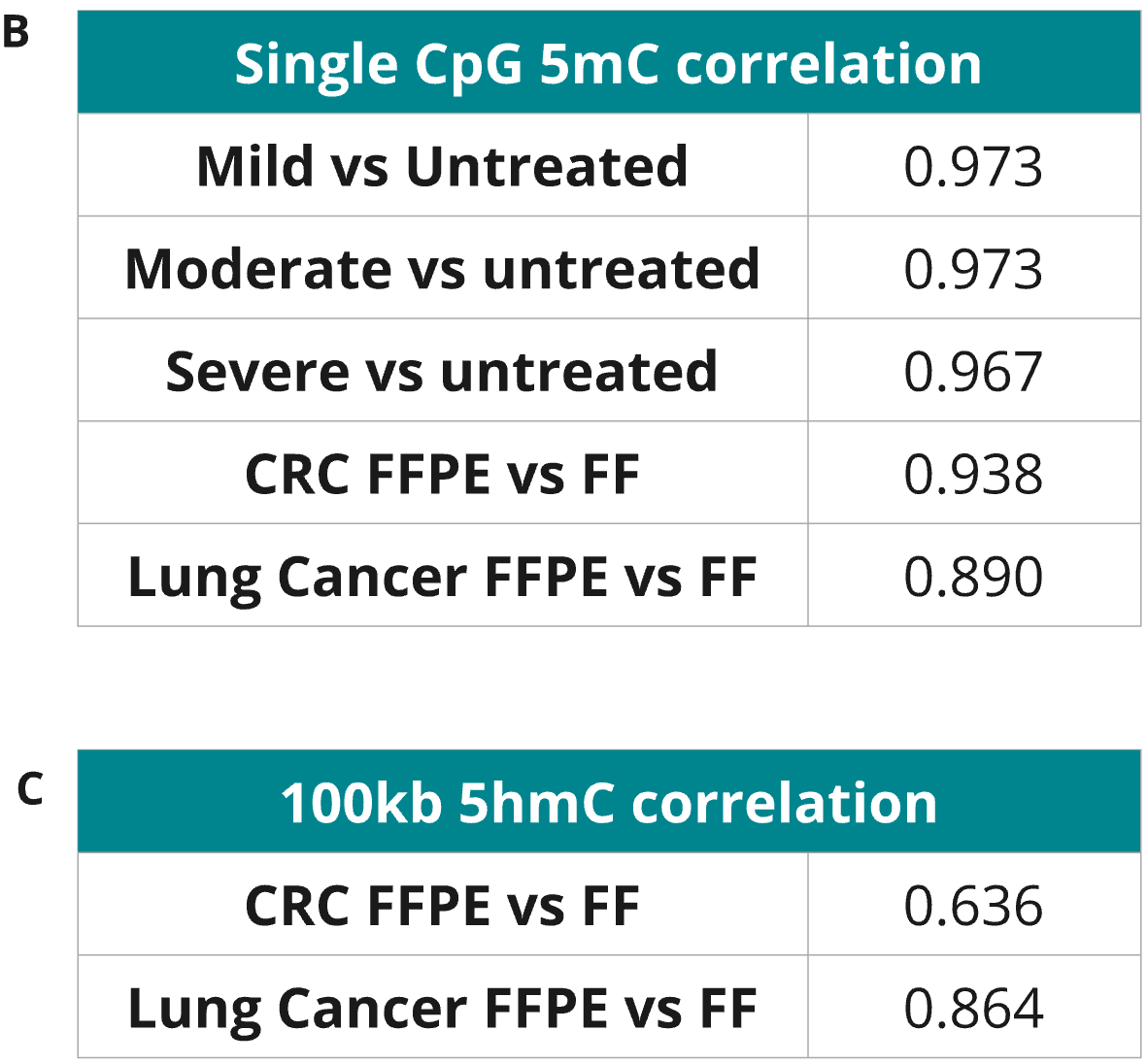
Correlation analysis of 6-base data from formalin-compromised DNA standards, Fresh Frozen (FF) and FFPE samples.
- A 5mC Pearson correlation was generated using the modality XPLR analysis software at single CpGs for duplicate libraries across fcDNA conditions. R² values >0.96 (B) demonstrate highly reproducible methylation calling between diverse samples even with severe damage, reducing background noise and increasing confidence in biological insights derived from 6-base FFPE data.
- Correlations for 5mC at single CpG level (B) and 5hmC across 100kb windows
- were also calculated clinical FF and FFPE samples also show very high 5mC correlation with R² of ~0.94 and ~0.89 for CRC and Lung cancer, respectively. Clinical FF and FFPE samples show robust 5hmC correlation with R² of ~0.64 and ~0.86 for CRC and Lung cancer, respectively, with the correlation partly drive by the lower prevalence of 5hmC in these samples (1.1-2.4%).
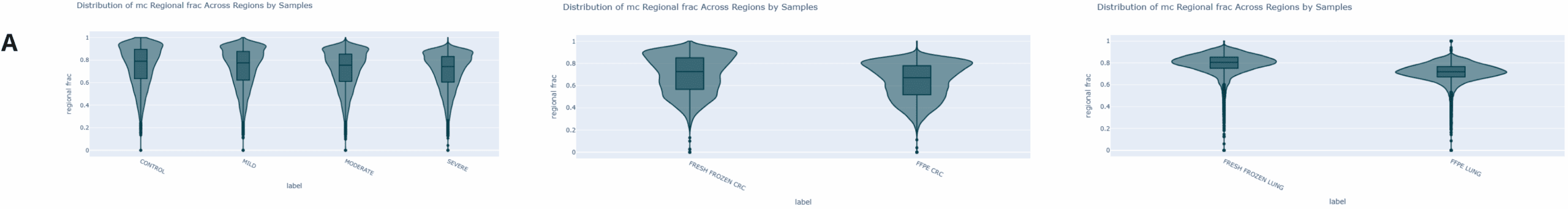
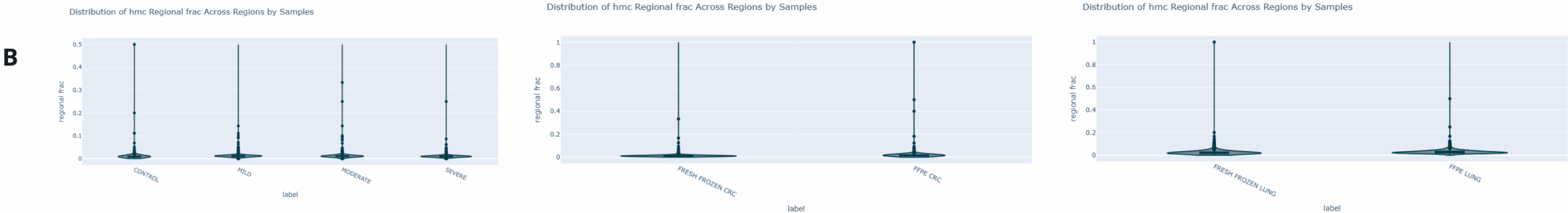
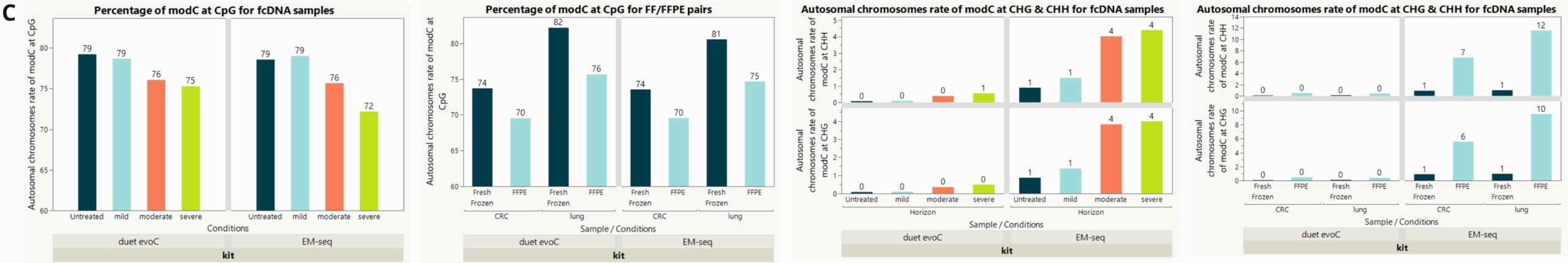
Genome-wide 5mC and 5hmC level analysis in CpG, CHG and CHH contexts.
- Violin plot generated using modality XPLR of the genome wide fraction of cytosines called as 5mC at CpGs for fcDNA and FF/FFPE samples, across 10kbp windows. 5mC levels decrease with increasing levels of formalin damage, with a ~4% drop for severe fcDNA relative to untreated control and a 5–8% reduction for FFPE compared to FF samples.
- Violin plot generated using modality XPLR of the genome wide fraction of cytosines called as 5hmC at CpGs for fcDNA and FF/FFPE samples, across 10kbp windows. 5hmC levels show a consistent ~1% increase with formalin damaged compared to untreated samples.
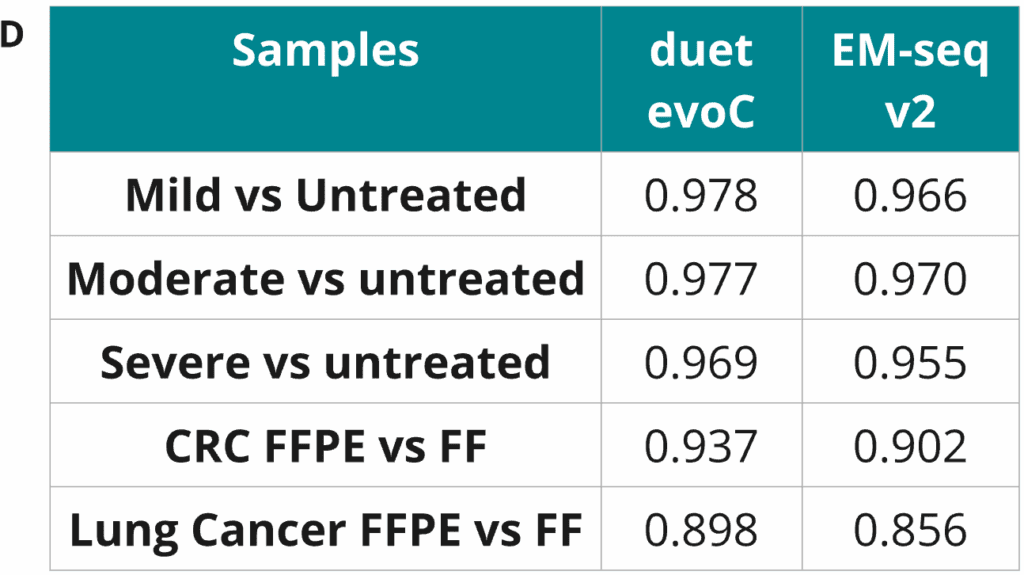
- Percentage of cytosines called as modC in CpG, CHG & CHH contexts across duet evoC and EM-seq v2 samples. A small decrease in modC levels associated with formalin damage and FFPE treatment is observed at CpGs. In contrast, a small increase in modC levels associated with formalin damage is observed at non-CpGs, especially for EM-seq. False methylation signals introduced by chemical damage can be corrected with duet evoC during the read-resolution step, supressing errors in non-CpG contexts. These data demonstrate EM-seq has a higher error rate and therefore worse signal to noise ratio, especially in non-CpG contexts, than duet evoC.
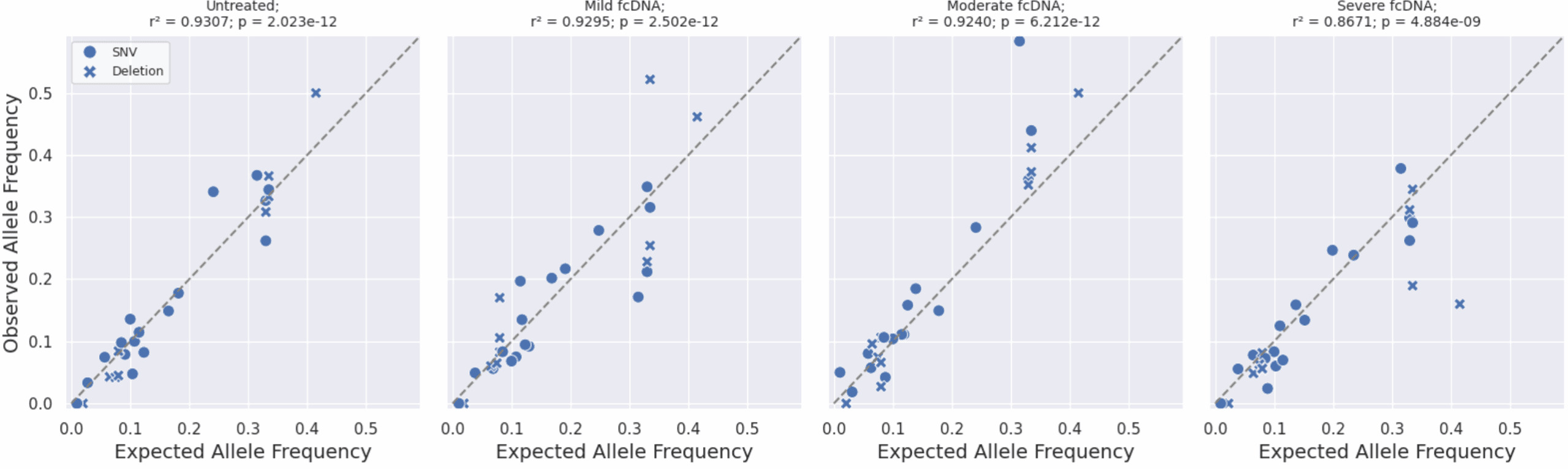
Observed vs Expected Variant Allele Frequencies (VAFs) on formalin-compromised DNA standards.
Expected allele frequencies were obtained from the manufacturer (Horizon Discovery) and observed allele frequencies were calculated by aligning reads to the reference genome and counting the number of reads supporting the variant as a fraction of the total coverage for that position. A high correlation with R² >0.92 up to moderate formalin damage and 0.86 for severe indicates that duet evoC supports accurate SNV calling from FFPE samples
We demonstrate compatibility of duet evoC with formalin-damaged DNA including clinical FFPE samples. Constructs made with formalin damaged samples retained highly accurate genetic and epigenetic information even at severe levels of damage. Very high 5mC correlation is observed between untreated and formalin-compromised DNA, between fresh-frozen (FF) and FFPE samples, and with minimal effects on overall GC bias for mild to moderately damaged DNA. Collapsing 6-base mC & hmC data to modC readout yielded higher correlations than EM-seq v2 and better agreement in overall modC calls across all contexts, in particular CHH/CHG, to untreated or FF DNA. Importantly, no significant effect was observed for allele frequency detection with formalin-induced damage. Overall duet evoC produces high quality genetic and epigenetic data with formalin damaged FFPE samples, opening up these challenging sample types to the advantages of 6-base analysis.
- Simultaneous sequencing of genetic and epigenetic bases in DNA, Füllgrabe and Gosal et al., Nature Biotechnology (2023) (duet multiomics solution technology paper)
- Quantitative Multiplex Reference Standard (Horizon Discovery)
- Hedegaard, Jakob et al. “Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue.” PloS one vol. 9,5 e98187. 30 May. 2014.



