Resources
Resource types
- Blog post
Understanding the factors that affect DNA methylation can open up new avenues to explore human health and disease.
- Blog post
5hmC is far more than just a transient step in the DNA demethylation pathway as it has broad implications for gene regulation, cellular differentiation, brain function, genome stability, and disease.
- Blog post
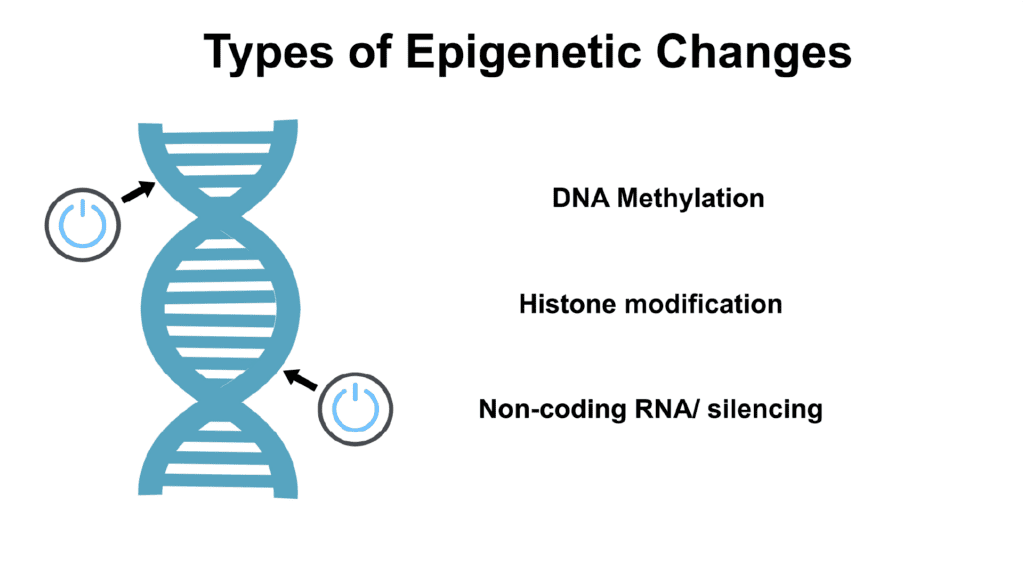
Explore the fascinating world of the different types of epigenetic modifications and the role of epigenetics in gene expression. Learn more.
- Blog post
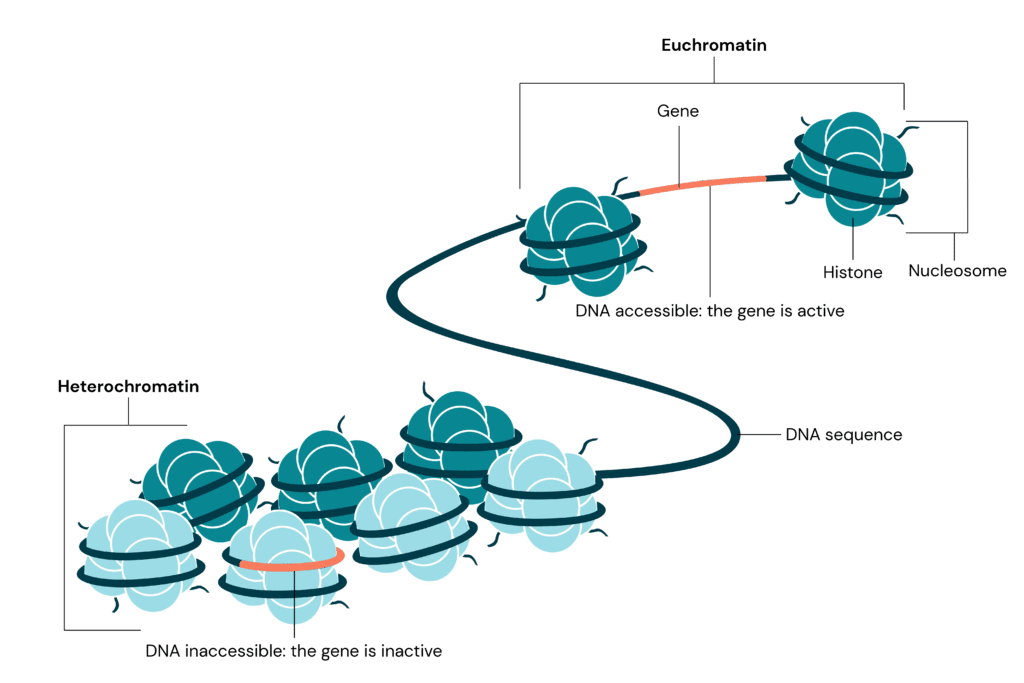
Heterochromatin is a condensed form of chromatin that plays an essential role in maintaining genomic stability. Disruptions in its formation can lead to disease. We explore the mysteries of heterochromatin and its vital role in genome organisation.
- Blog post
Understanding how alterations in CpG island methylation contribute to disease states, including cancer, may provide novel targets for treatment.
- Webinar
Watch this presentation, given by Chennan Li, PhD, from the National Cancer Institute, at the American Association of Cancer Researchers annual meeting 2025.
- Case study
The Munich Leukaemia Laboratory (MLL) are investigating the role of the 6-base genome to develop workfows that enable the detection of epigenetic patterns in leukemia patients.
- Poster
The combination of PARPi (Talazoparib – TAL) and DNMTi (Decitabine – DAC) functions synergistically to inhibit PCa cell lines and patient-derived PTOs.
- Product sheet
Ihr duet multiomics solution evoC-Produktblatt finden Sie hier
- Poster
Circulating tumor DNA (ctDNA) purity can be used to predict therapy response of metastatic castration-resistant prostate cancer (mCRPC) to PD-L1/PARP1 inhibition.
- Webinar
By employing the 6-base genome via duet evoC, we have identified differentially methylated genes that may be responsible for therapy resistance and tumor aggression in pancreatic cancer.