Cells contain all the genetic information required for protein synthesis for a biological system to function, and the development of high-throughput DNA sequencing has been key in elucidating cellular processes and understanding how variation and mutations play a role in disease states, such as cancer.
But it’s not just genetic sequences that control cellular fate and function – epigenetic modifications regulate gene expression in response to changes in behaviour or environment and play a key role in many biological processes such as development and ageing, as well as disease.
Variation in an individual’s genetic sequence will be associated with variation in DNA methylation that can functionally determine predisposition for disease.
Measuring both genetic and epigenetic variation in the context of the other is therefore important for elucidating dynamic biological processes, allowing a more comprehensive view of cellular function.
The challenge of combined genetic and epigenetic sequencing
However, capturing both genetic and epigenetic information simultaneously comes with significant challenges due to the limitations of current sequencing technology.
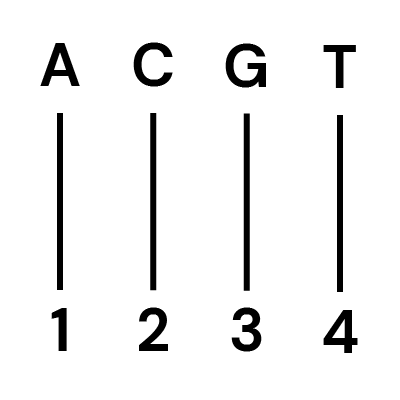
Next generation sequencing technologies only capture four letters or information states in their readout, the canonical bases A, T, C, and G.
Base conversion chemistries can be used to differentiate unmodified Cytosine from its most prevalent epigenetic variants, 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), but this means sacrificing the accurate identification of one of the canonical bases (typically the base Thymine) and missing important genetic sequence information (Figure 1).
| State | Standard sequencing protocol | Protocol with C→T deamination |
|---|---|---|
| 1 | A | A |
| 2 | C/mC/hmC | mC/hmC |
| 3 | G | G |
| 4 | T | C/T |
So, when looking to identify 5mC or 5hmC, the unmodified C base is converted to U (and so is read as a T in sequencing) therefore compromising the detection of C-to-T changes in the genetic sequence, which are the most common mutations in mammalian genomes and cancer.
C-to-T changes due to base conversion also cause ambiguity in genomic alignment, resulting in slower, more expensive, and less accurate read mapping.
To date, sequencing both genetic and epigenetic information from a single sample has come with a significant time and cost burden, requiring separate sequencing workflows, increased sample requirements and inexact integration of the data, wherein phase information is lost.
biomodal’s duet multiomics solution +modC for improved biological characterisation
To overcome these challenges of combined genetic and epigenetic analysis, we developed duet multiomics solution +modC, a whole genome methodology that can capture all four canonical bases as well as modified C in a single workflow.
Our platform consists of a pre-sequencing enzymatic workflow that uses standard molecular biology techniques to prepare the sample for sequencing, as well as decoding software to resolve the raw sequencing data.
We have optimised this workflow for Illumina sequencers, but other sequencing adapters can be substituted, meaning the workflow can be used with any sequencing platform capable of decoding at least four genetic bases.
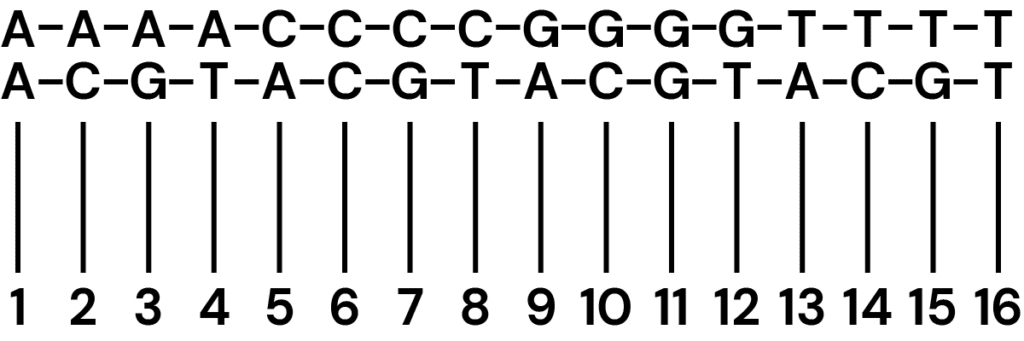
Two-base coding system for combined genetic & epigenetic sequencing
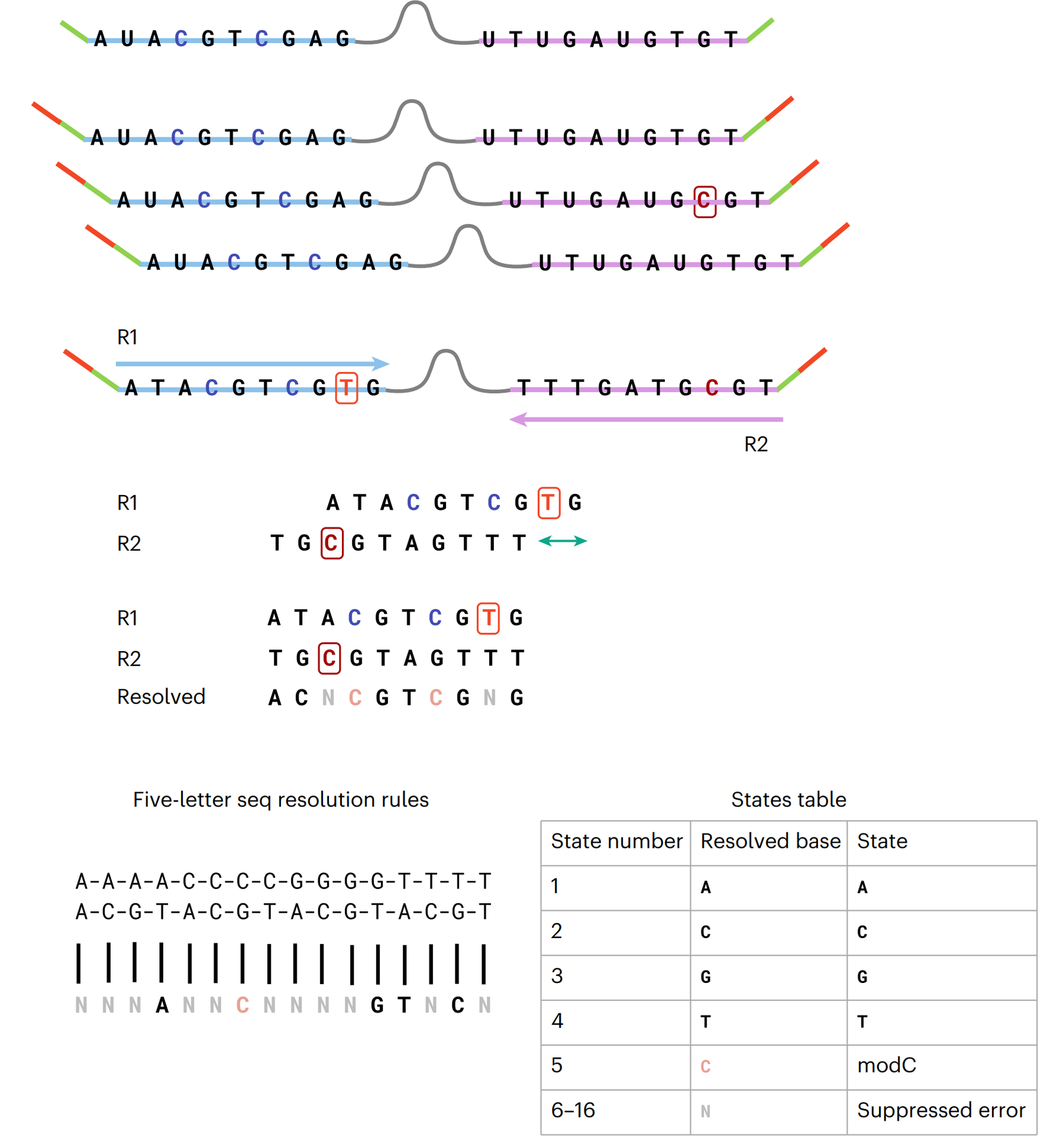
Standard sequencing results in a four-state (or four-letter) readout – here, we use a two-base coding approach where the combination of two bases denotes the original state, enabling up to 16 states to be decoded (Figure 2).
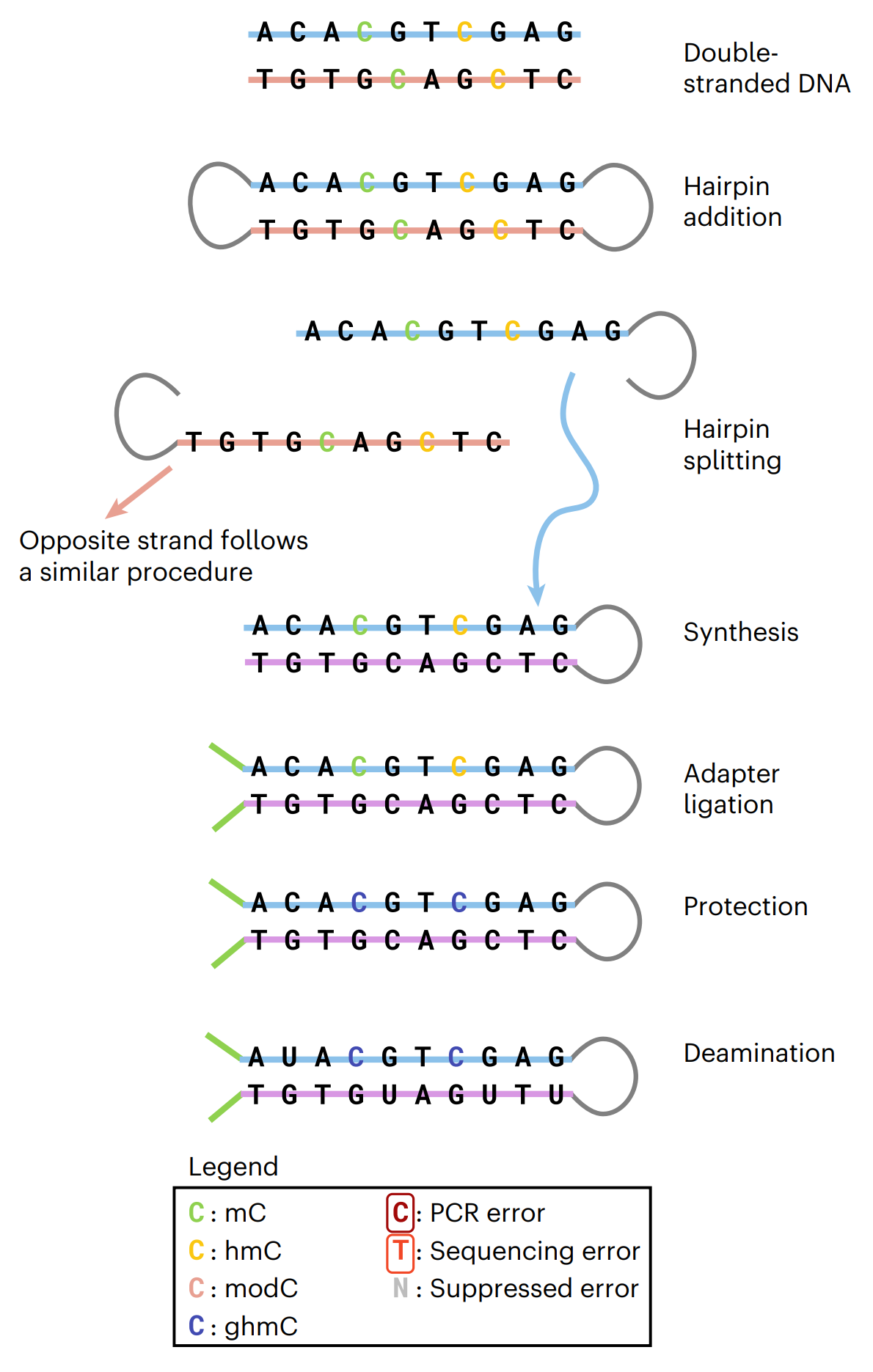
The workflow begins with the ligation of sample DNA fragments to short, synthetic hairpin adapters at both ends.
Each strand is then copied by DNA polymerase to form a construct with the original sample strand connected to its complementary copy strand via a synthetic hairpin.
Sequencing adapters are ligated at each end. Modified Cs are enzymatically protected by oxidation and glycosylation, unmodified Cs are then deaminated to uracil.
The base converted DNA template can be amplified by PCR and is then ready for sequencing (Figure 3).
Our bioinformatic decoding software then performs pairwise alignment of the original and complementary strands and computationally resolves the sequencing data, producing a resolved base that corresponds to a single genetic or epigenetic letter.
Any errors arising from sample prep, amplification, or incorrect base-calling during sequencing will occur independently to cognate bases on each strand and so will result in an impossible base pair combination which is filtered out in subsequent analysis, providing inherent error suppression capability (Figure 4).
We performed 5-Letter sequencing on a mixed B-lymphoblast cell line and generated both genetic and epigenetic data in a single sequence.
Having all four genetic states in the read sequences allowed for standard genomic alignment, significantly reducing execution times compared to WGBS and EM-seq.
Comparison of data quality to WGBS and EM-seq showed increased specificity and sensitivity for the detection of epigenetic marks, and higher accuracy of read mapping.
Simultaneous detection of both genetics and epigenetics in cis on the same DNA molecule also allowed us to detect differential DNA methylation between alleles.
Variant-associated methylation (VAM) and methylation quantitative trait loci (methQTLs) can be used to identify regulatory sequence variation that underpin many diseases.
Meeting the low input demands of liquid biopsy samples
The combination of both genetic and epigenetic information on a single DNA fragment can provide key insights on the dynamic interactions that are occurring, offering significant advantages in many research areas such as cellular fate studies, stem cell differentiation, population genomics, and cancer biology.
One important area of interest is liquid biopsy for tumour diagnosis and disease monitoring, as combining DNA methylation information with the genetic sequence in cell-free DNA (cfDNA) from blood has shown significantly increased sensitivity to detect tumour DNA.
We applied duet +modC to a cfDNA sample from a human cancer patient with gastric cancer and achieved a very high-quality sequencing data on a sample containing only 2ng DNA.
This shows that our method can detect both genetic and epigenetic data from the same low input sample, and so could therefore overcome the challenges of working with valuable and low input biological samples for diagnostics.
Future developments
The 5hmC modification is a known marker of disease states, including early cancer detection.
The two-based coding nature of our platform means we were able to further adapt the platform to differentiate between 5mC and 5hmC.
Our system also has the potential to measure additional epigenetic modifications, such as formylcytosine, methyladenine, or carboxycytosine.
We have shown that 5-Letter sequencing can deliver accurate, genetic and epigenetic sequences on the same DNA molecule, in a single workflow, that is faster and more accurate than bisulfite sequencing or EM-seq.
Our workflow allows both genetics and epigenetics to be studied in the context of the other, offering comprehensive biological insight with ease of use and reduced overhead.
The duet multiomics solution +modC platform can also be used to generate accurate data from valuable, low input biological samples or cell-free DNA which could feasibly transform cfDNA analysis and liquid biopsy for detection of early cancer.