biomodal
- Robert Crawford
- Jamie Scotcher
- Fabio Puddu
- Dan Brudzewsky
- Jane D Hayward
- Andrada Tomoni
- David Morley
- Páidí Creed
- Philippa Burns
- Joanna D Holbrook
DNA comprises molecular information stored in genetic and epigenetic bases, both of which are vital to our understanding of biology. duet multiomics solution +modC is a technology which sequences at base resolution the complete genetic sequence integrated with modified cytosine (modC). duet multiomics solution +modC has previously been demonstrated using genomic and cell free DNA. Here we demonstrate its performance relative to formalin-compromised DNA standards [1] (fcDNA) as well as DNA extracted from formalin-fixed and paraffin embedded (FFPE) samples of two colorectal cancer (CRC) patients.
FFPE samples represent an important resource for studying genetic and epigenetic information from archived tissues. However, DNA damage induced by formalin fixation (e.g. deamination, fragmentation or nucleic acid cross-linking) can lead to decreased data quality from next generation sequencing (NGS) relative to ‘gold standard’ fresh-frozen samples. Damage may manifest in lower library yields and insert sizes as well as higher duplication and lower coverage rates[2].
When studying formalin-damaged DNA with duet multiomics solution +modC we found expected changes in yield and insert size, as well as C:G>T:A mutations. Background modC levels decreased ~5% in CpG contexts and increased ~0.5% in CHH/CHG contexts between untreated and severe formalin damaged (DIN ≤ 2.0) standards. Importantly, no substantial differences in variant allele frequency (VAF) were observed for a set of reference variants, even at severe formalin damage. Overall, duet multiomics solution +modC is compatible with formalin damaged samples such as FFPE.
Key outputs
- BAM containing resolved reads with modC annotations
- VCF containing germline or somatic variants
- CytosineReport file containing modified Cytosine levels for each CpG
duet multomics solution + modC applied to formalin-damaged DNA.
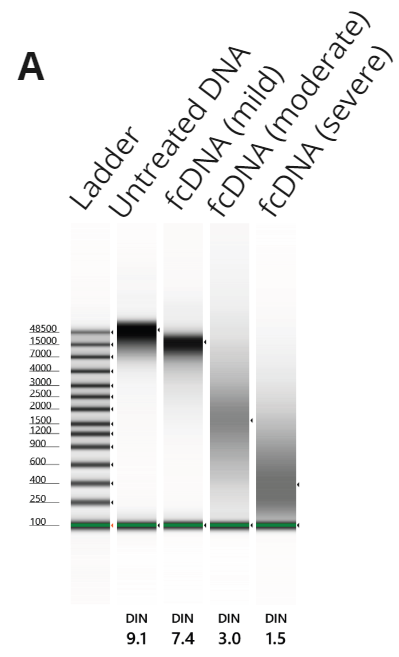
To determine if duet multomics solution + modC is compatible with formalin-damaged genomic DNA, we used formalin-compromised Quantitative Multiplex Reference Standard (QMRS) DNA (fcDNA) from Horizon Discovery alongside the QMRS untreated control. fcDNA damage levels were mild, moderate and severe corresponding to DNA Integrity Number (DIN) ranges of ≥ 5.1, 2.5 – 5.0 and ≤ 2.0.
- Tapestation traces for untreated QMRS gDNA, QMRS fCDNA at mild, moderate and severe formalin damage. DIN scores are shown below each trace.
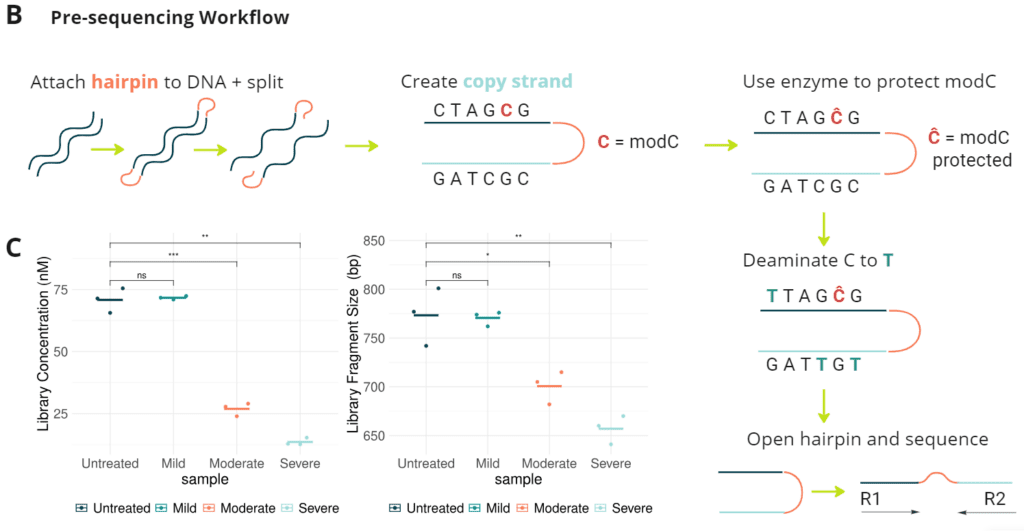
- Pre-sequencing Workflow. Triplicates of 40 ng sheared DNA are end-repaired, A-tailed and ligated to hairpins. Top and bottom strands are separated, then a copy of the original strand is synthesized. Modified cytosines (modC e.g. 5mC; 5hmC) are protected enzymatically before the final deamination step. The resulting construct contains a two-base code discriminating five bases (A, C, T, G, modC) and eleven error codes.
- Final library concentration and insert size for QMRS gDNA (untreated) and QMRS fcDNA (mild, moderate, severe). Increasing formalin damage reduced library yield and insert size.
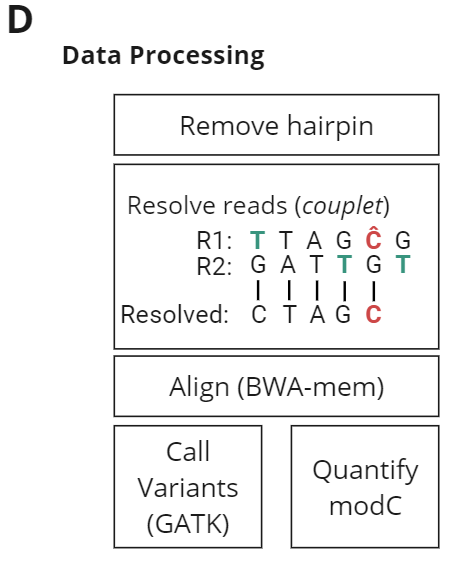
- Data Processing. After Illumina PE150 sequencing, data was processed through biomodal‘s pipeline for the analysis of +modC libraries, to remove hairpins, resolve the modification state of each cytosine by comparing Read1 and Read2, and align resolved reads to the reference genome.
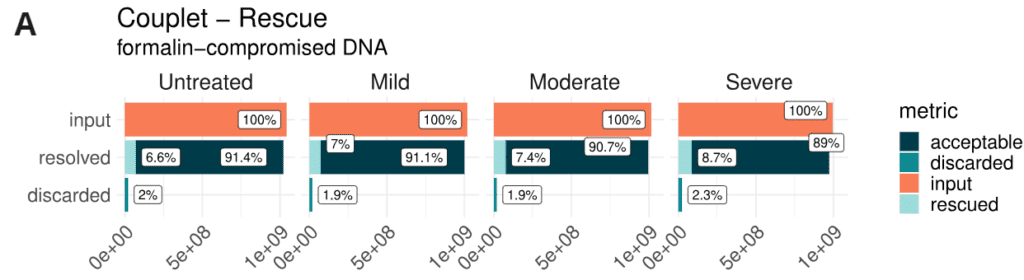
- Couplet resolution and rescue metrics. Read pairs received by couplet are classified as acceptable, when they can be resolved without re-alignment, or rescued, when re-alignment is necessary to achieve resolution. Read pairs are marked as discarded when they cannot be resolved. The highest level of formalin damage only results in a small increase in the number of discarded or non-acceptable reads, while most non-acceptable reads are rescued during resolution.
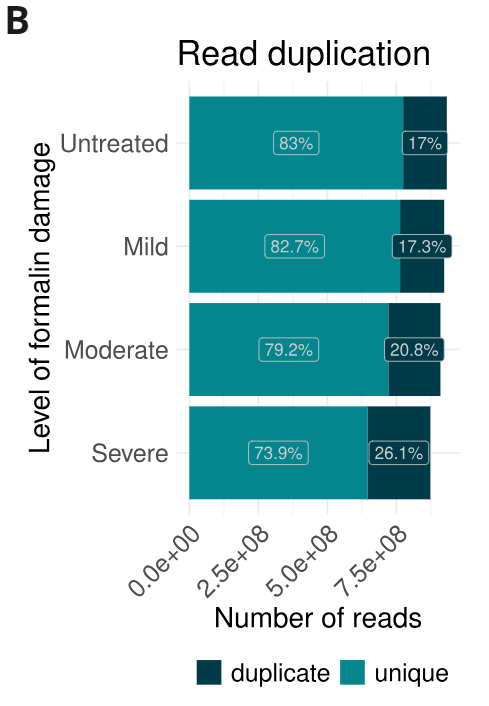
- Formalin damage leads to an expected increase in read duplication because of a reduction in library complexity.
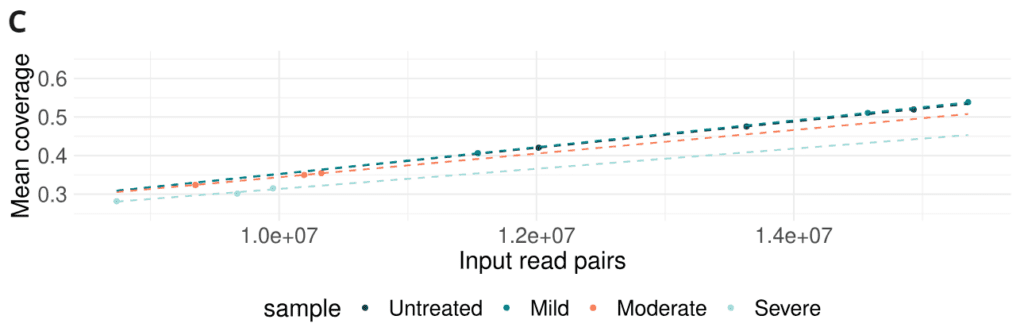
- Combined effects of formalin damage lead to a small reduction in sequencing yield (mean coverage achievable per number of input read pairs).
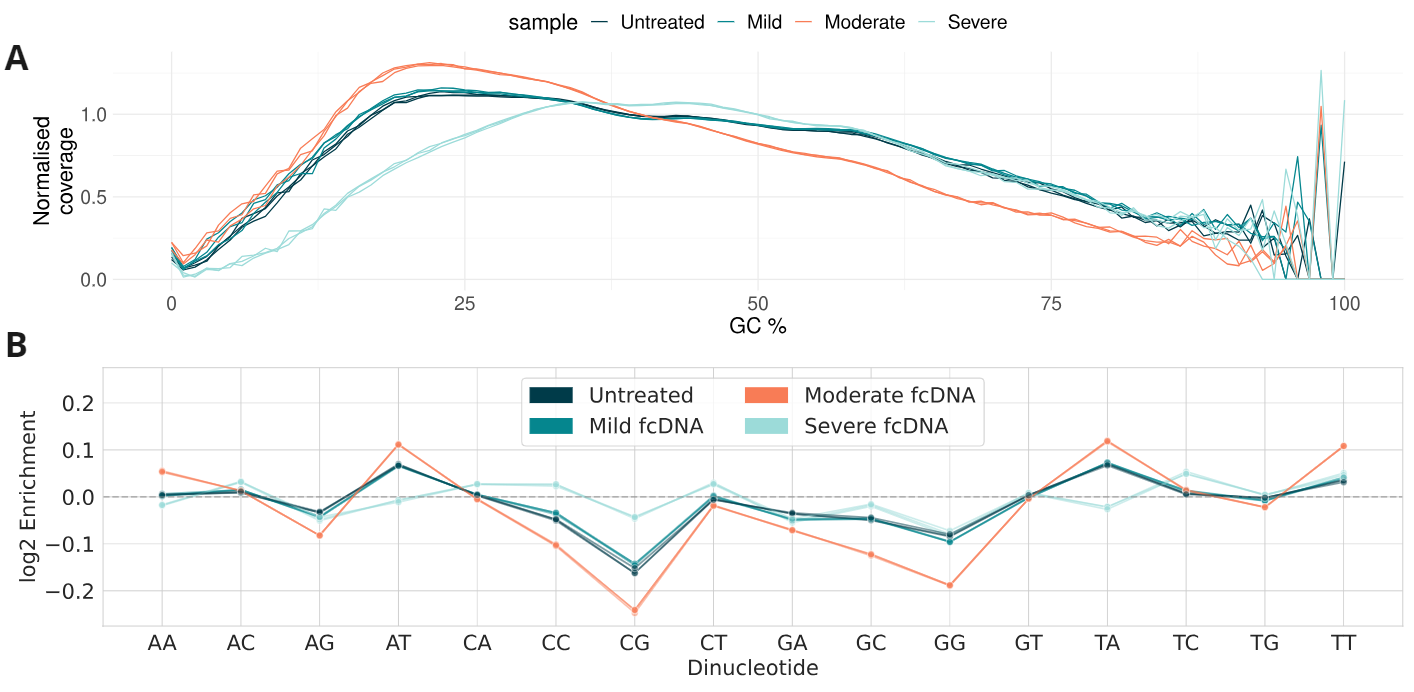
Analysis of nucleotide coverage biases in formalin-compromised DNA standards.
- Formalin damage only mildly alters GC coverage bias. Three technical replicates are shown as separate lines.
- Log2 enrichment of dinucleotide coverage in samples over their respective frequencies in the GRCh38 reference genome; formalin damage only mildly alters dinucleotide coverage bias.
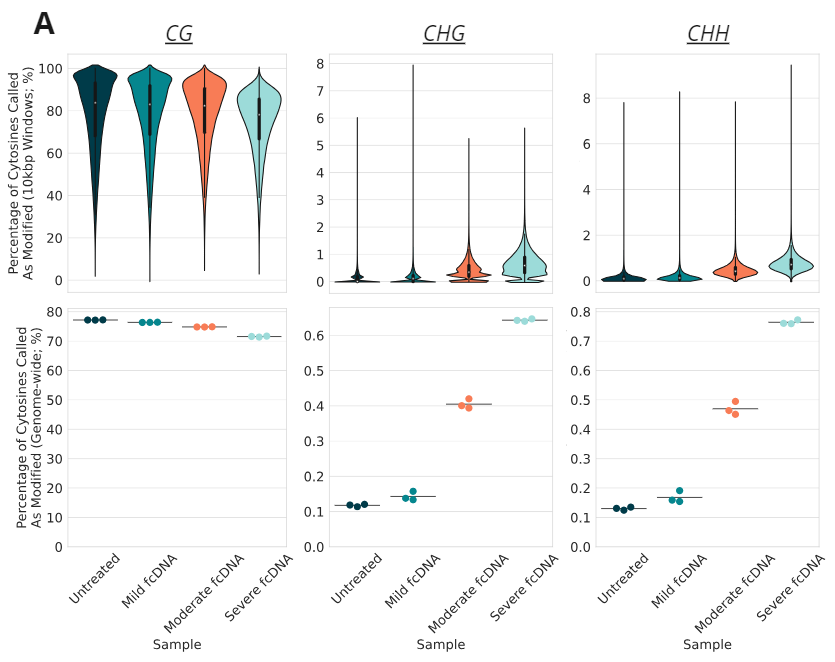
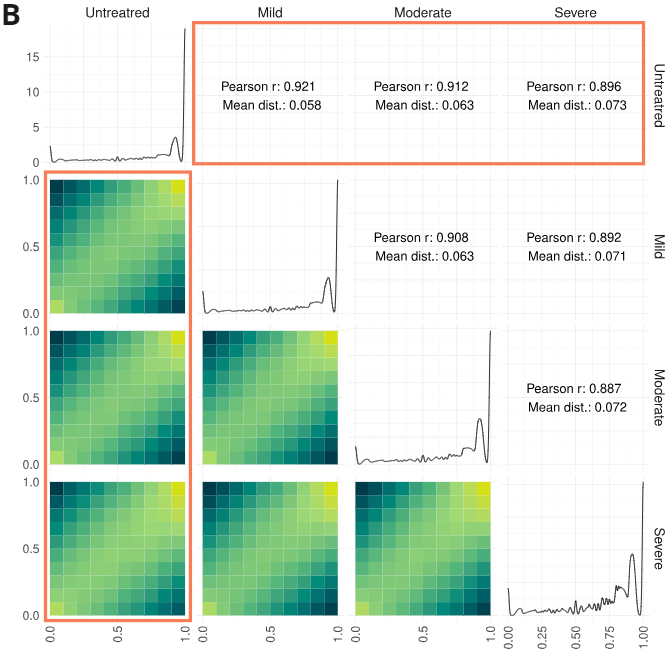
Epigenetic analysis of biomodal's pipeline run on formalin-compromised DNA standards.
- Percentage of cytosines called as modC in CG, CHG & CHH contexts, calculated for 10kbp windows and genome-wide (top and bottom rows, respectively).
- Correlation of CpG methylation levels across undamaged samples or samples with different levels of formalin damage. Bottom quadrant: each CpG was been binned in 10% bins depending on its methylation state as measured in different samples; the heatmap shows the log10-scaled density of CpGs in each bin. Top quadrant: Pearson r, and mean distance of each CpG from the diagonal are reported for all the comparisons shown in the bottom quadrant. Diagonal: density plot of CpG methylation levels in each of the four samples analysed.
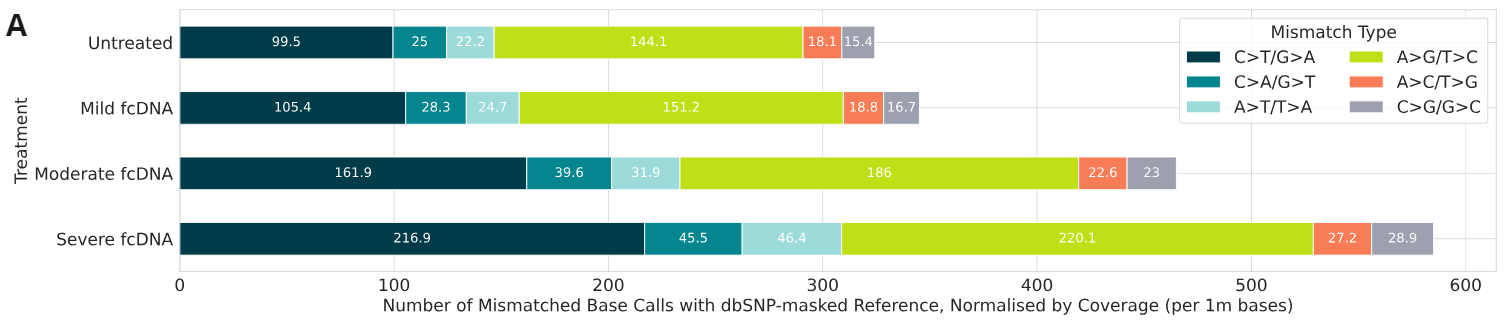
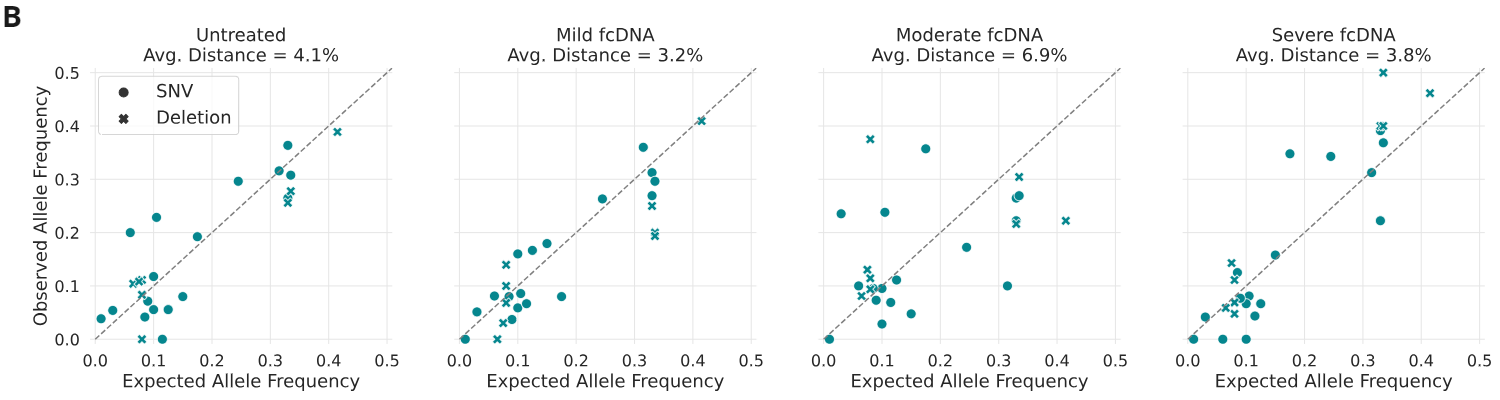
Analysis of base calls and variant allele frequencies (VAFs)
- Mismatched bases between formalin-compromised DNA standards and GRCh38 reference genome; common SNPs identified in dbSNP were masked from the reference. Mismatch counts were normalised by sample coverage and are represented per 1 million base calls. Note C>T/G>A and A>G/T>C mutations are not suppressed by Couplet‘s error suppression.
- Observed vs Expected Variant Allele Frequencies; expected allele frequencies were obtained from Horizon(1) and observed allele frequencies were calculated by aligning reads to the reference genome and counting the number of reads supporting the variant as a fraction of the total coverage for that position.
NA12878: undamaged gDNA
FFPE1: mild-moderate damage (DIN=5.0)
FFPE2: moderate-severe damage (DIN=3.2)
duet multiomics solution +modC applied to FFPE samples
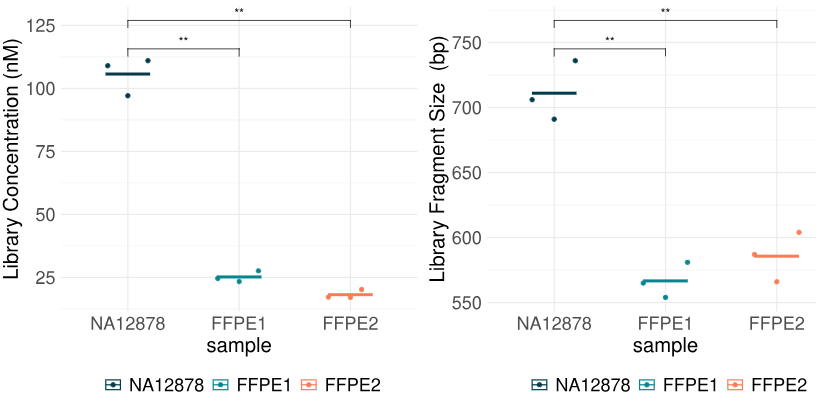
To determine if duet multiomics solution +modC is compatible with formalin-fixed paraffin-embedded samples, we used as input DNA extracted from FFPE samples derived from two colorectal cancer (CRC) patients. DNA extraction was performed using the chemagic truXTRAC DNA FFPE Kit. FFPE1: Female, CRC Grade 1, Stage IIIB, age 70-85, DIN = 5.0. FFPE2: Male, CRC Grade 1, Stage I, age 70-85, DIN = 3.2. Processing extracted DNA followed the same pre-sequencing workflow and data processing described in Panels 1 B & D. Here, 80ng of sheared extracted DNA from each FFPE sample were processed in triplicate. As with fcDNA lower yield and insert size was observed for formalin-damaged samples relative to undamaged NA12878 gDNA controls.
Analysis of metrics produced by biomodal's pipeline run on FFPE samples
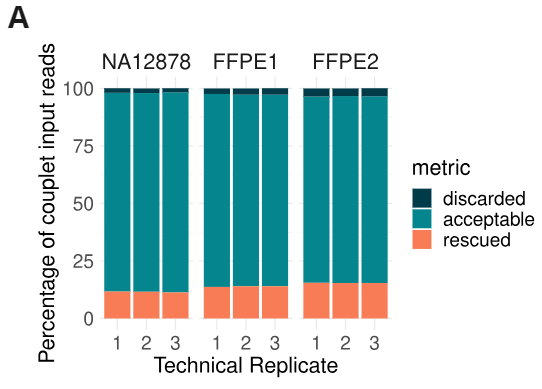
- Couplet resolution and rescue metrics. As described in Figure 2, read pairs received by couplet are classified as acceptable, rescued, or discarded. As observed for fcDNA samples, the highest level of formalin damage (FFPE2) only results in a small increase in the number of discarded reads, while most non-acceptable reads are rescued during resolution. Three technical replicates are shown for each sample.
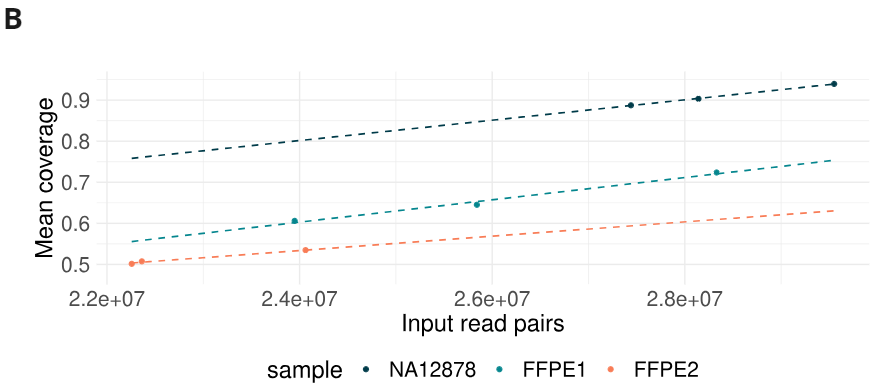
- Formalin damage lead to a reduction in sequencing yield (mean coverage achievable per number of input read pairs).
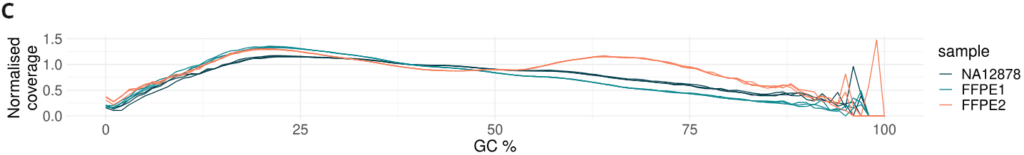
- Formalin damage only mildly alters GC coverage bias. Three technical replicates are shown as separate lines.
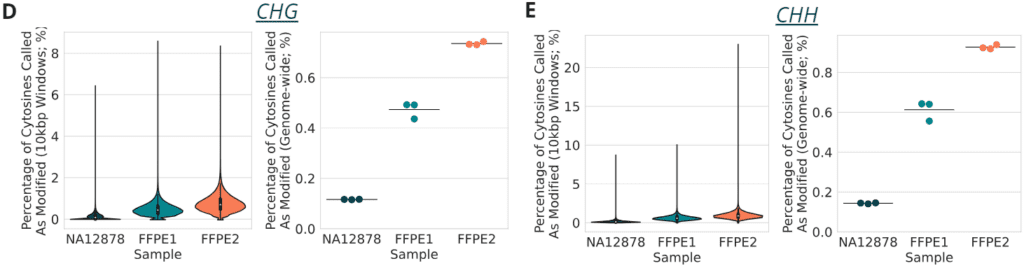
- and E: percentage of cytosines called as modC in CHG and CHH contexts, respectively; genome-wide modC percentages are shown as well as per 10kbp window.
- each point represents a technical replicate and mean values for samples are shown as lines. FFPE samples were derived from two colorectal cancer (CRC) patients; FFPE1: Female, CRC Grade 1, Stage IIIB, age 70-85, DIN = 5.0; FFPE2: Male, CRC Grade 1, Stage I, age 70-85, DIN = 3.2.
In conclusion we demonstrate the compatibility of duet multiomics solution +modC with formalin-damaged DNA. While damage resulted in lower library yields and insert sizes relative to undamaged controls, high accuracy genetic and epigenetic base resolution data is produced even at severe levels of formalin-induced damage. No effect on allele frequency detection was observed for the fcDNA QMRS standards. We note small changes in modC calling dependent on sequence context: a decrease for CpG, and an increase for CHH/CHG. We hypothesise this may be due to slightly different formalin-induced deamination pathways acting on unmodified cytosines (predominately found at CHH/CHG) and modified cytosines (much more prevalent in CpG contexts).
- Quantitative Multiplex Reference Standard (Horizon Discovery)
- Hedegaard, Jakob et al. “Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue.” PloS one vol. 9,5 e98187. 30 May. 2014.




