- Jack M Monahan
- Aurel Negrea
- Lisabet Andreasen
- Paula Golder
- David J Morley
- Rosie Telford Spencer
- Minna Taipale
- Jens Fullgrabe
- Walraj S. Gosal
- Páidí Creed
The use of cell-free DNA (cfDNA) as a non-invasive liquid biopsy facilitates the monitoring and early detection of genetic and epigenetic changes. The typical cfDNA yield from a phlebotomy is 5-10ng/ml, corresponding to 1,500 to 3,000 copies of the haploid genome. Targeted sequencing enables the high-resolution measurement of variation in a subset of the genome. Sequencing to a comparable depth with a limited DNA input can be impractical for a whole-genome approach.
duet multiomics solution evoC duet multiomics solution evoC is a new sequencing technology that derives all four genetic bases alongside the ability to distinguish the modification status at the fifth carbon of cytosines, discriminating 5-Methylcytosine (5mC) from 5-Hydroxymethylcytosine (5hmC) (6-Base calling), in a single experiment. These epigenetic modifications comprise one fundamental pathway by which mammalian genes can be transcriptionally silenced or activated.
Here we demonstrate that duet evoC from low input cfDNA samples, targeted with a single hybrid capture is sufficient to obtain accurate genetics and epigenetics for regions of interest. These phased data avoid the otherwise complex data integration that is necessary with alternative epigenetic NGS techniques.

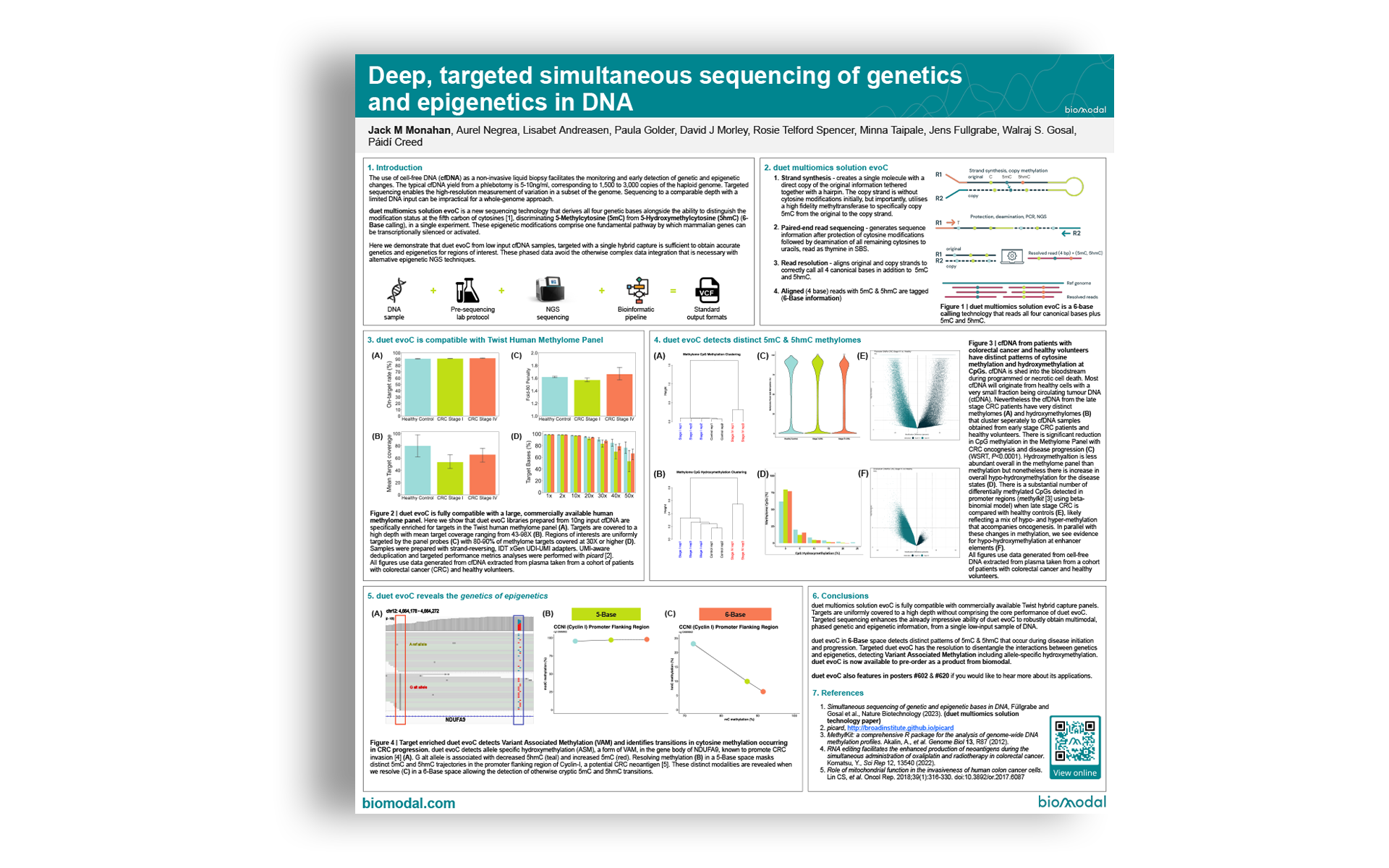
- Strand synthesis – creates a single molecule with a direct copy of the original information tethered together with a hairpin. The copy strand is without cytosine modifications initially, but importantly, utilises a high fidelity methyltransferase to specifically copy 5mC from the original to the copy strand.
- Paired-end read sequencing – generates sequence information after protection of cytosine modifications followed by deamination of all remaining cytosines to uracils, read as thymine in SBS.
- Read resolution – aligns original and copy strands to correctly call all 4 canonical bases in addition to 5mC and 5hmC.
- Aligned (4 base) reads with 5mC & 5hmC are tagged (6-Base information)
duet multiomics solution evoC is a 6-base calling technology that reads all four canonical bases plus 5mC and 5hmC.
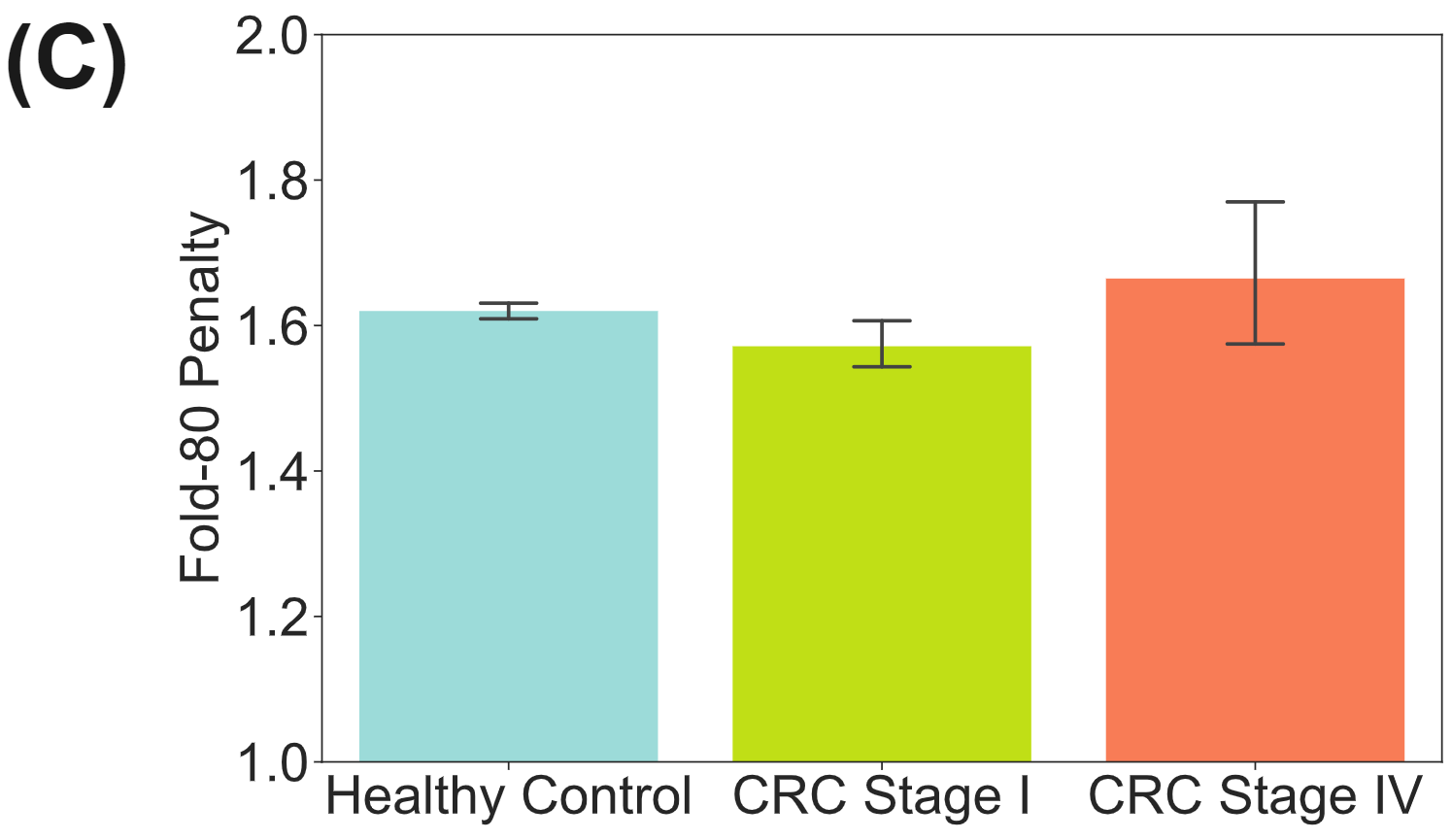
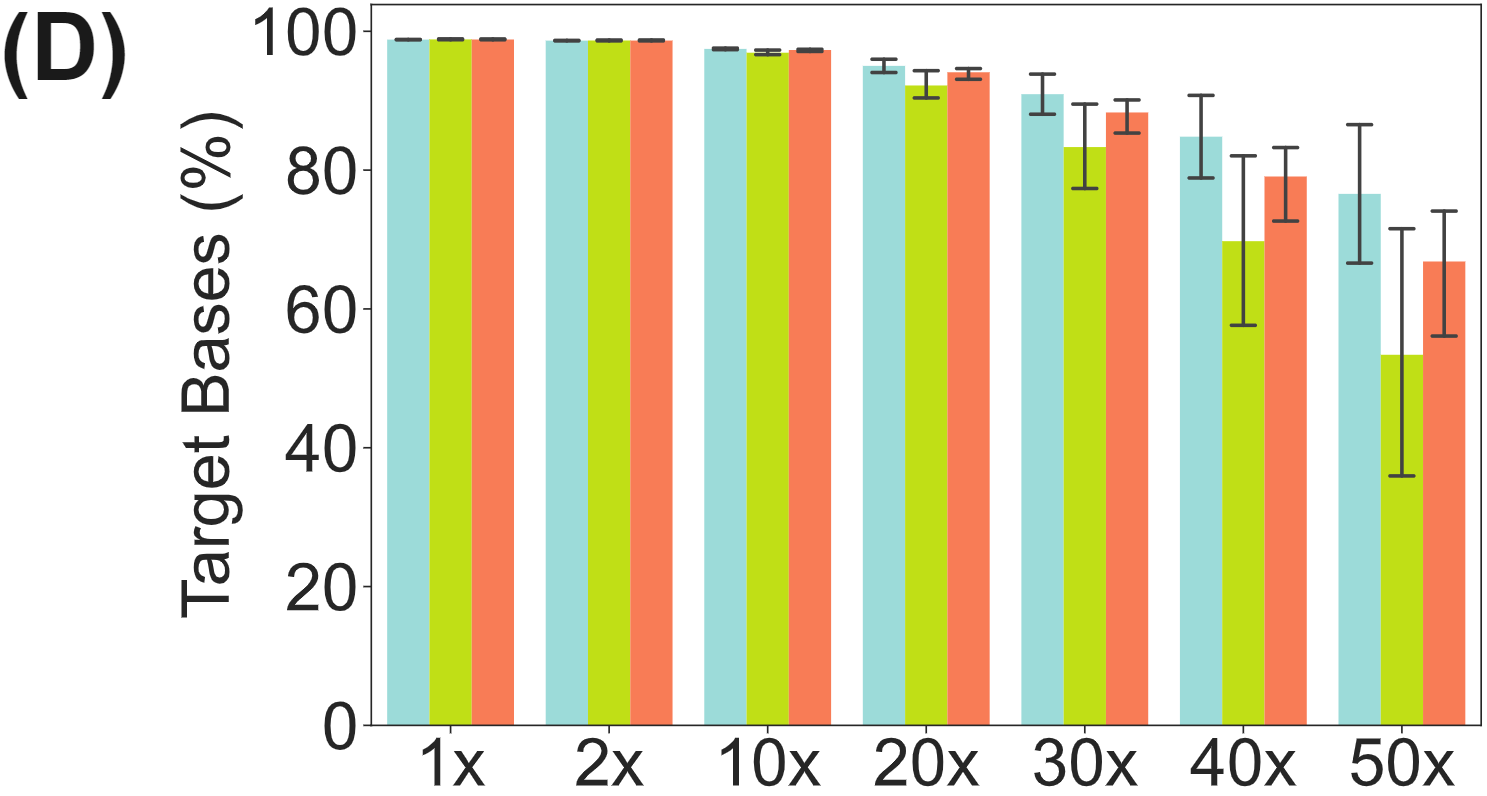
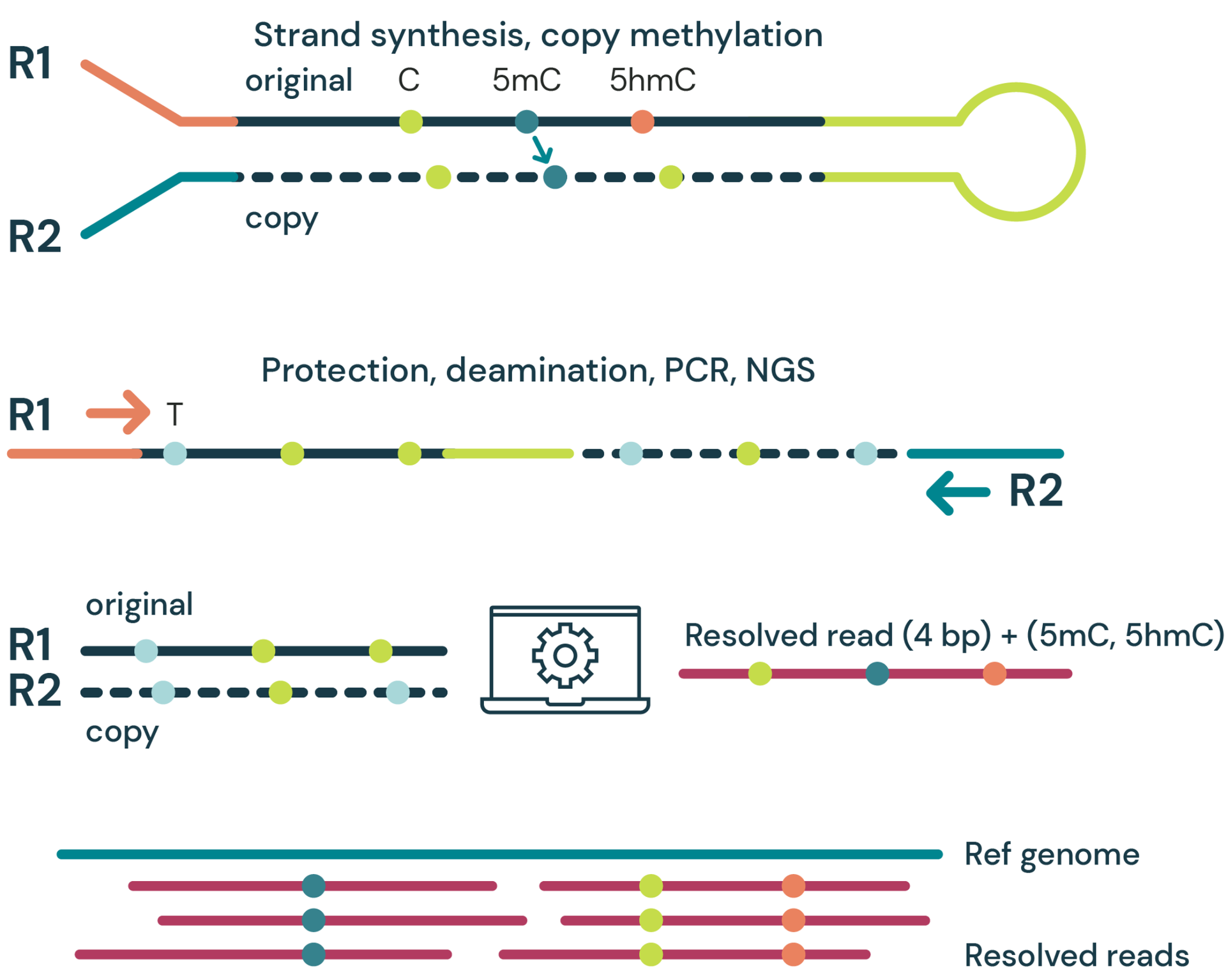
Figure 2 | duet evoC is fully compatible with a large, commercially available human methylome panel.
Here we show that duet evoC libraries prepared from 10ng input cfDNA are specifically enriched for targets in the Twist human methylome panel (A).
Targets are covered to a high depth with mean target coverage ranging from 43-98X (B).
Regions of interests are uniformly targeted by the panel probes (C) with 80-90% of methylome targets covered at 30X or higher (D).
Samples were prepared with strand-reversing, IDT xGen UDI-UMI adapters. UMI-aware deduplication and targeted performance metrics analyses were performed with picard.
All figures use data generated from cfDNA extracted from plasma taken from a cohort of patients with colorectal cancer (CRC) and healthy volunteers.
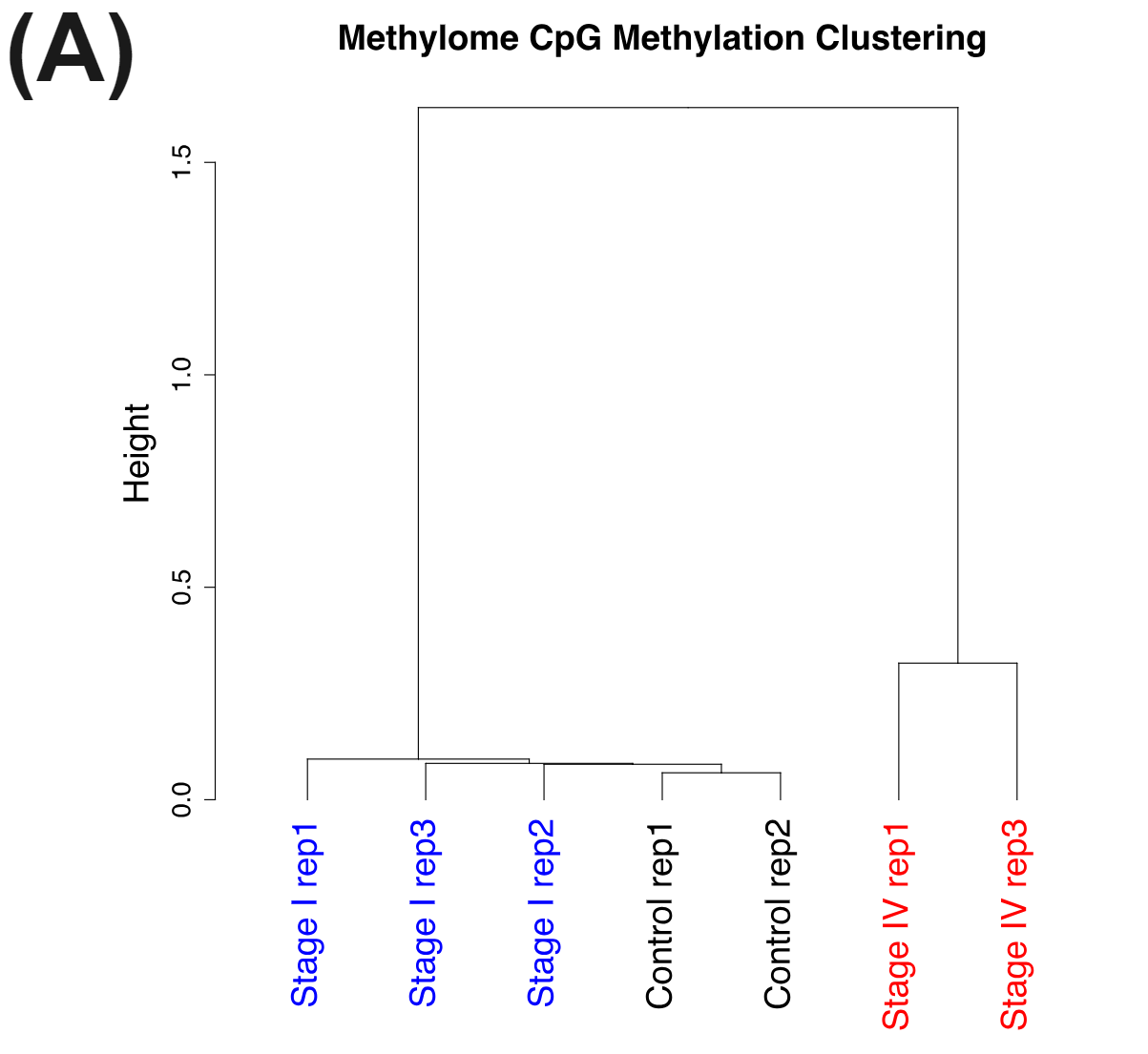
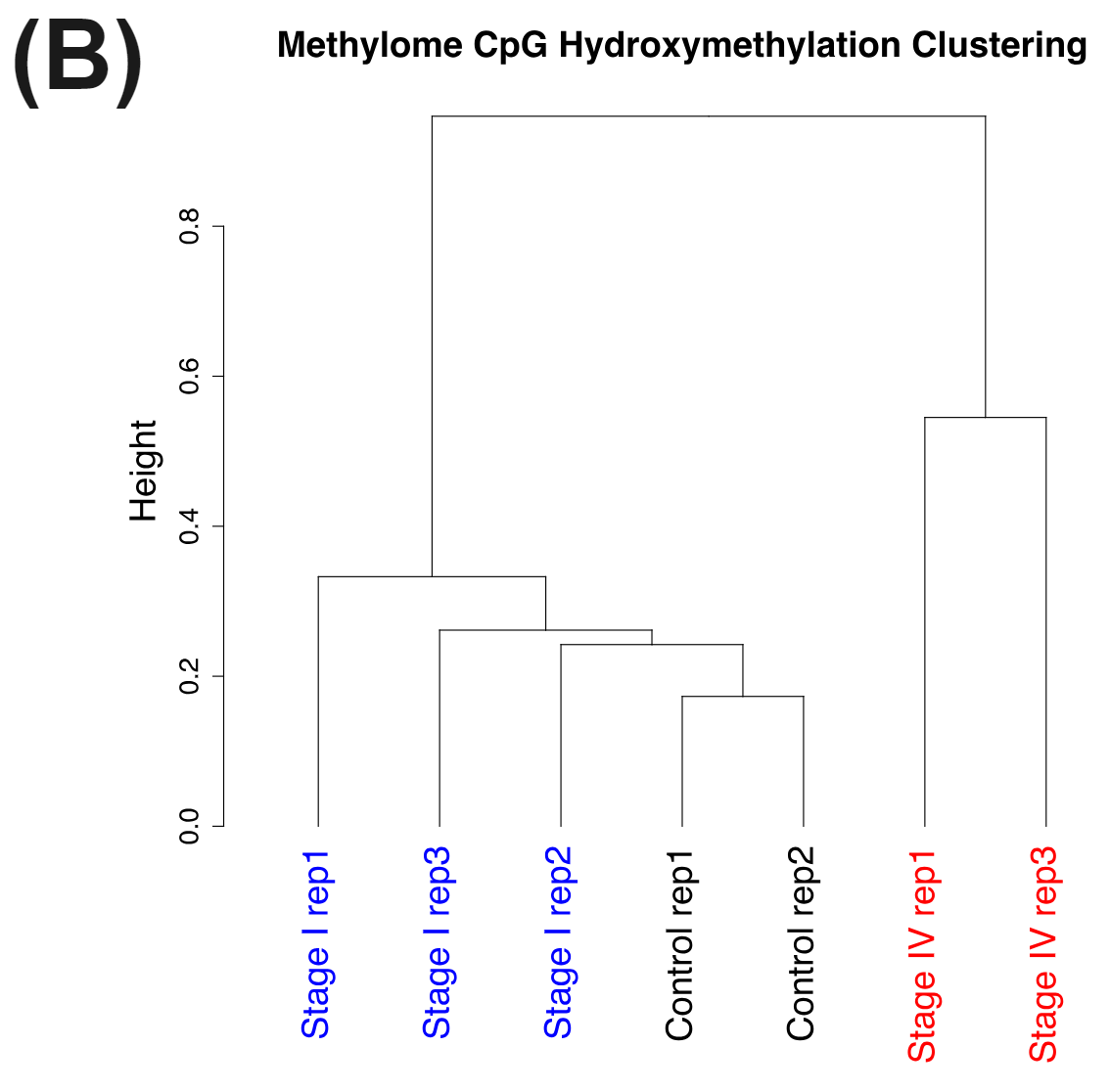
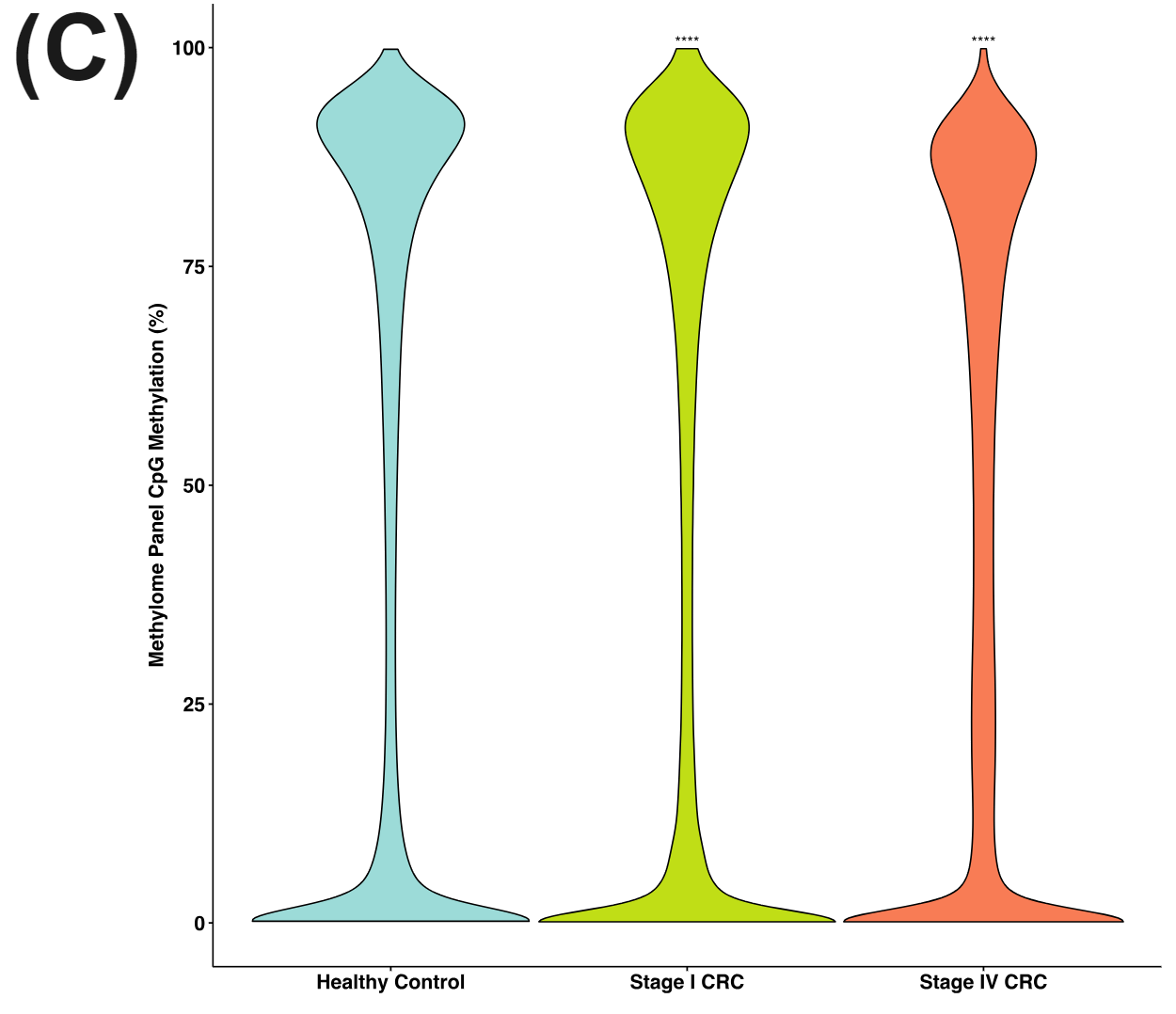
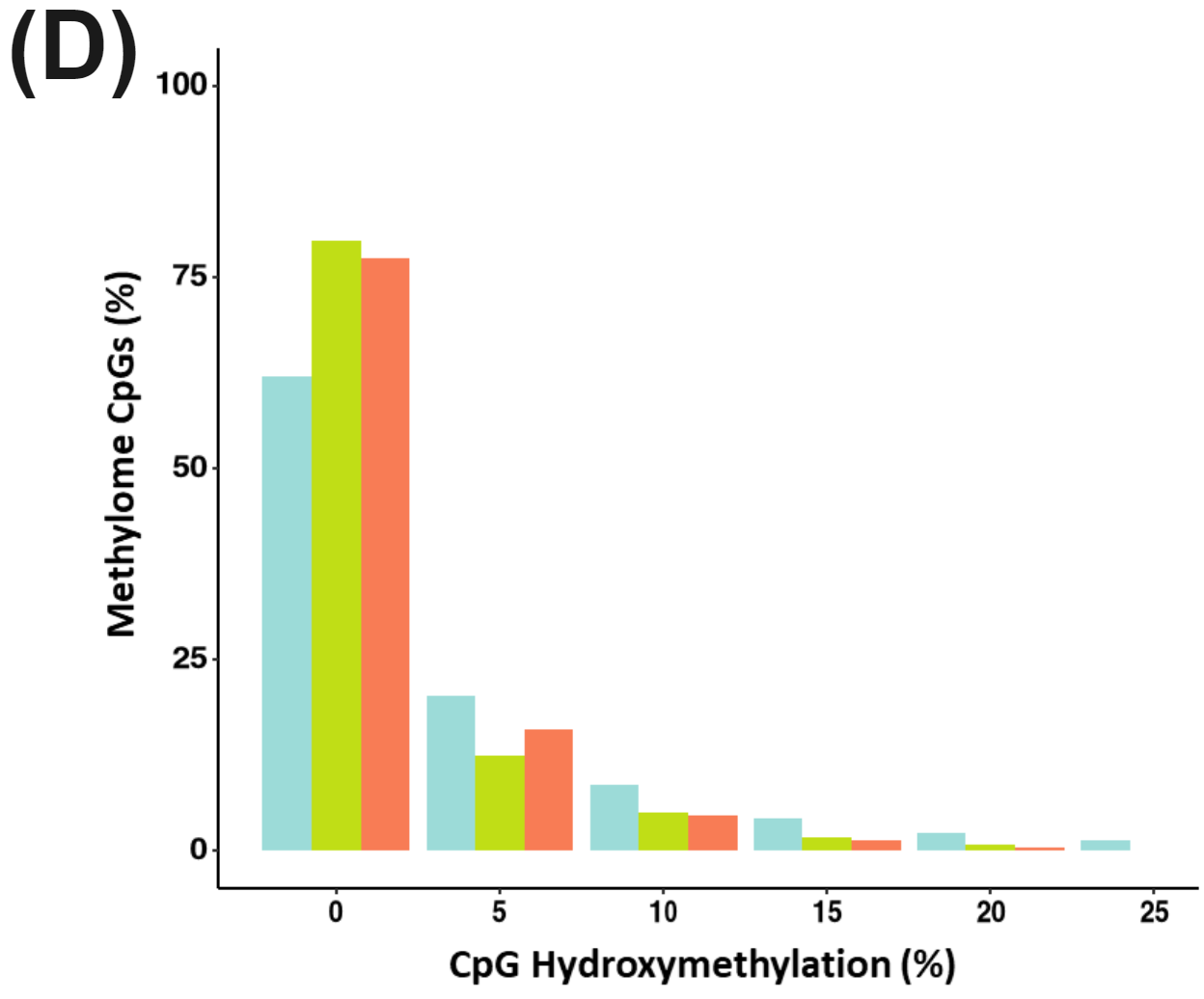
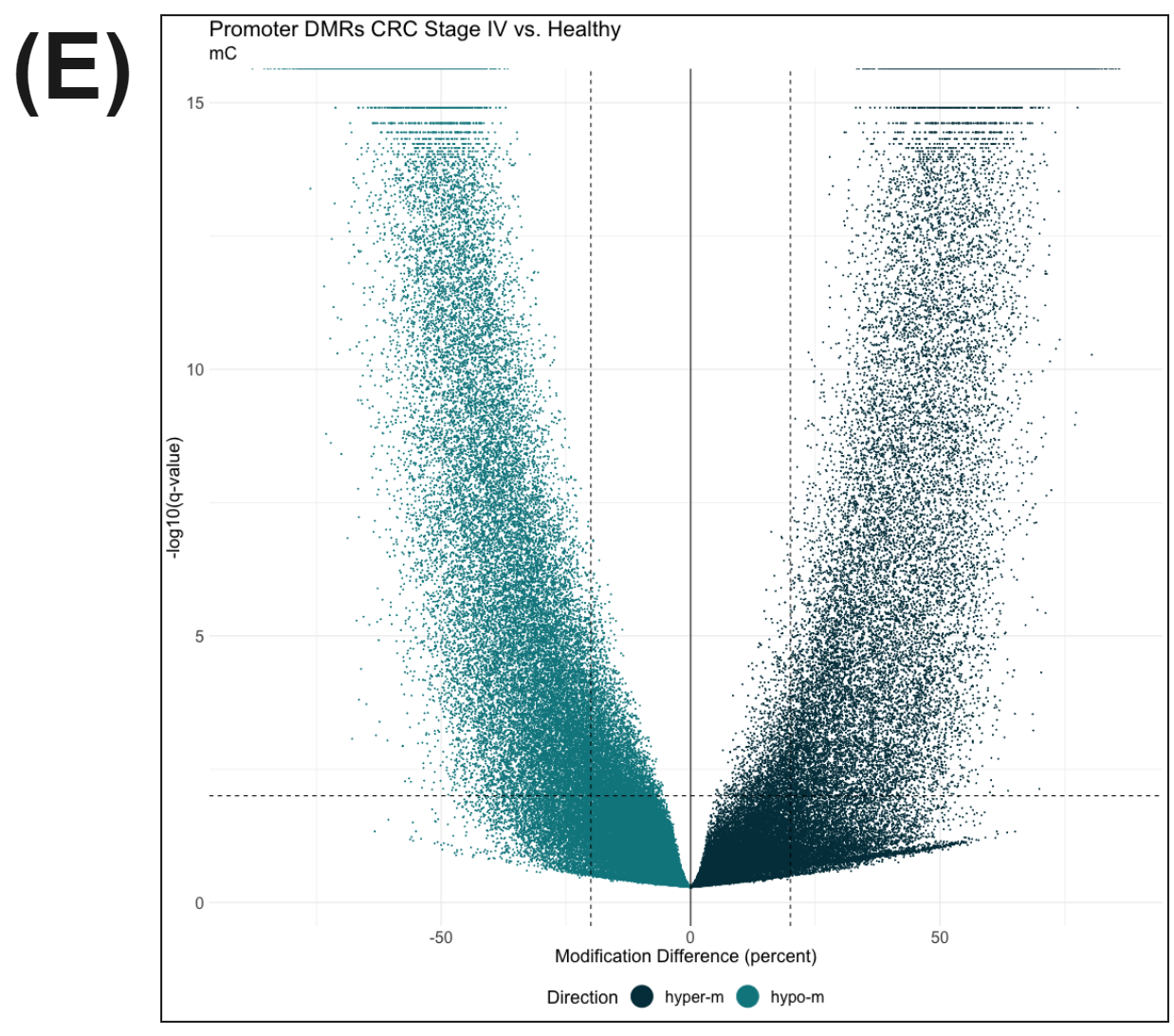
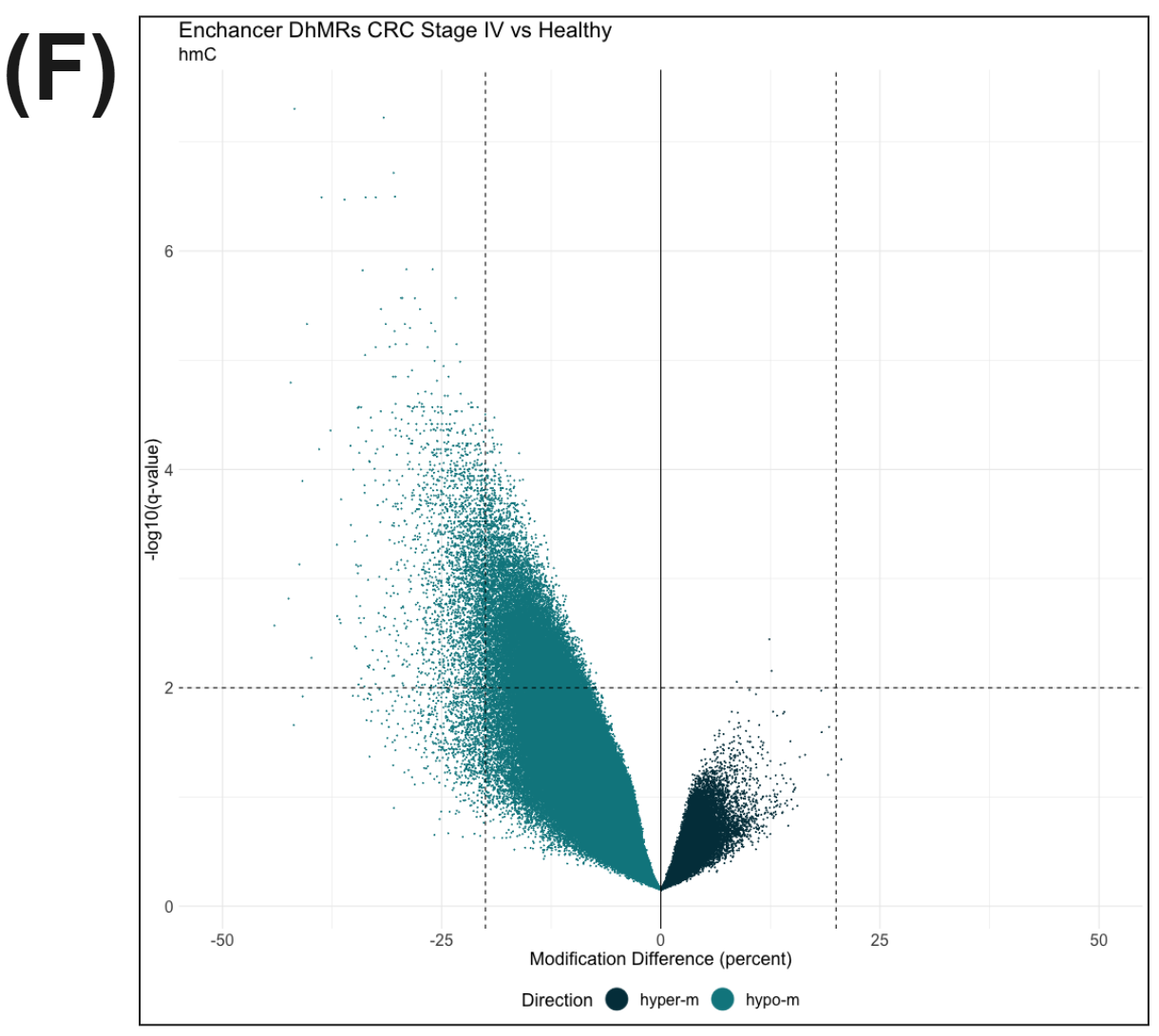
Figure 3 | cfDNA from patients with colorectal cancer and healthy volunteers have distinct patterns of cytosine methylation and hydroxymethylation at CpGs.
cfDNA is shed into the bloodstream during programmed or necrotic cell death. Most cfDNA will originate from healthy cells with a very small fraction being circulating tumour DNA (ctDNA). Nevertheless the cfDNA from the late stage CRC patients have very distinct methylomes (A) and hydroxymethylomes (B) that cluster seperately to cfDNA samples obtained from early stage CRC patients and healthy volunteers.
There is significant reduction in CpG methylation in the Methylome Panel with CRC oncognesis and disease progression (C) (WSRT, P<0.0001). Hydroxymethyaltion is less abundant overall in the methylome panel than methylation but nonetheless there is increase in overall hypo-hydroxymethylation for the disease states (D).
There is a substantial number of differentially methylated CpGs detected in promoter regions (methylkit using beta-binomial model) when late stage CRC is compared with healthy controls (E), likely reflecting a mix of hypo- and hyper-methylation that accompanies oncogenesis.
In parallel with these changes in methylation, we see evidence for hypo-hydroxymethylation at enhancer elements (F).
All figures use data generated from cell-free DNA extracted from plasma taken from a cohort of patients with colorectal cancer and healthy volunteers.
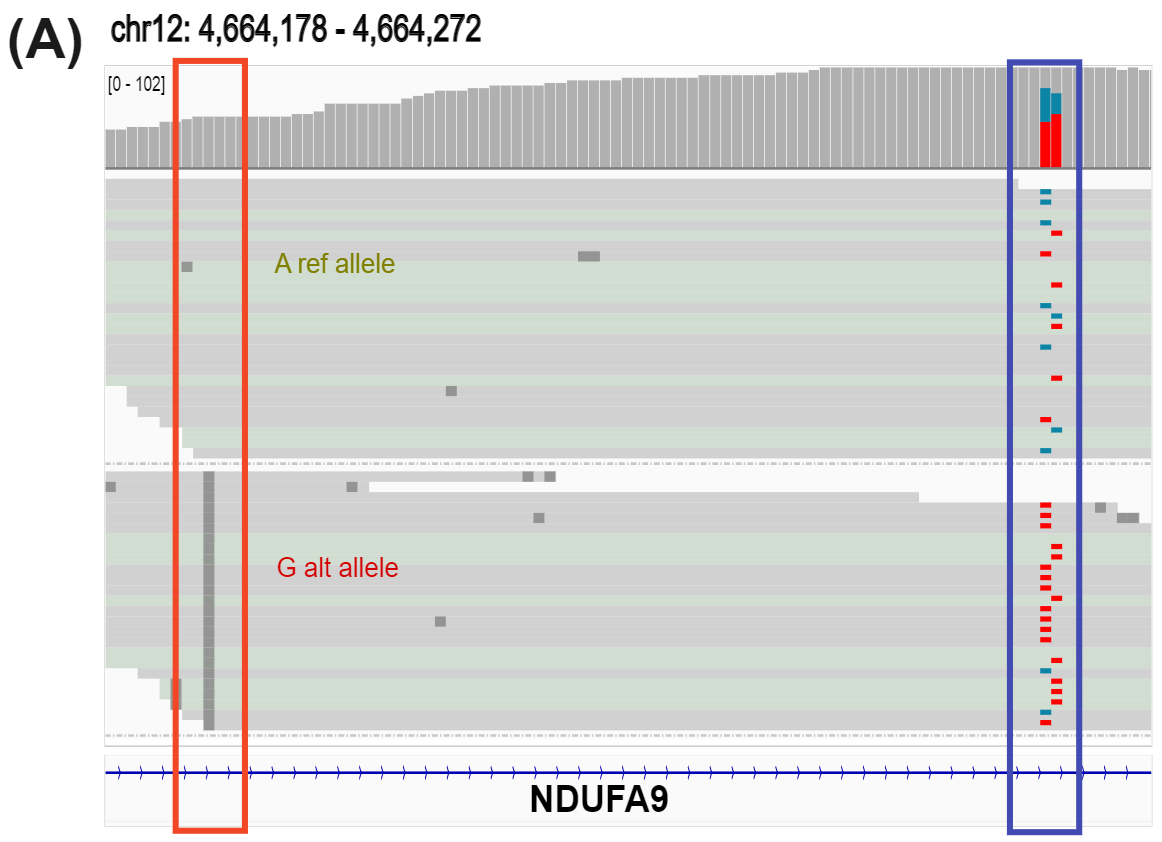
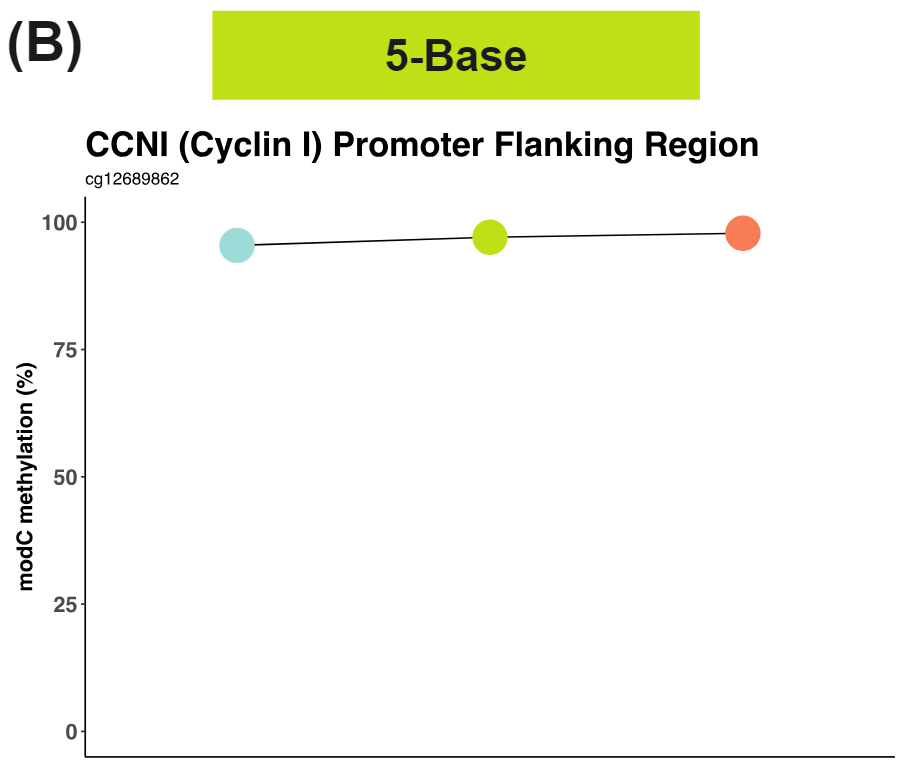
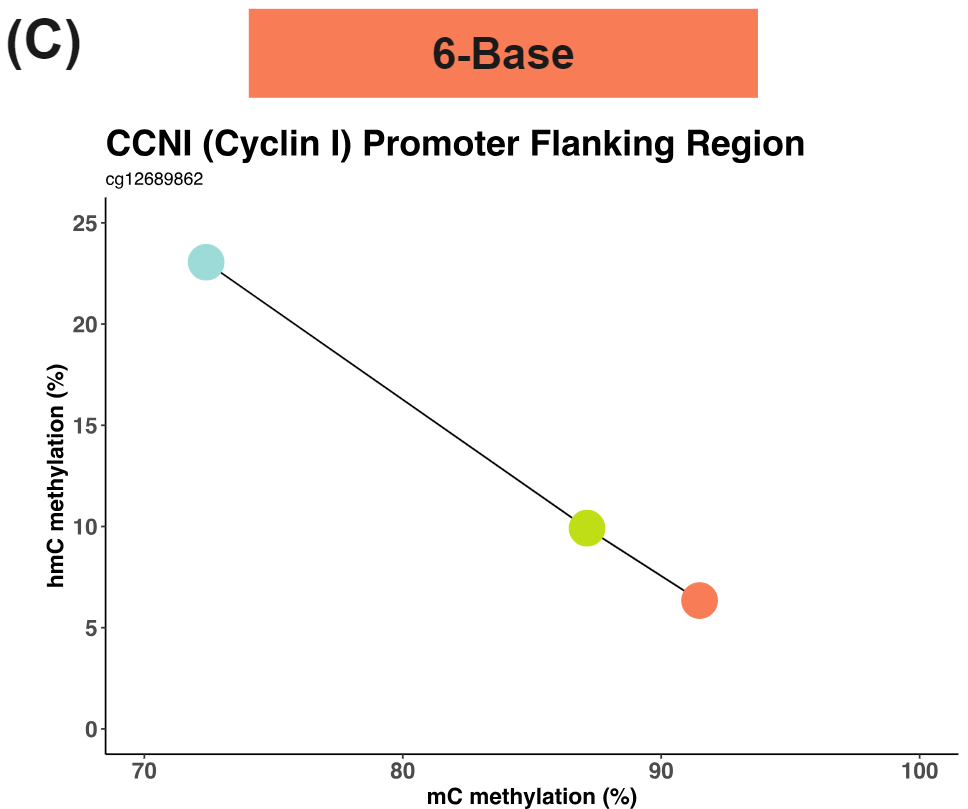
Figure 4 | Target enriched duet evoC detects Variant Associated Methylation (VAM) and identifies transitions in cytosine methylation occurring in CRC progression.
duet evoC detects allele specific hydroxymethylation (ASM), a form of VAM, in the gene body of NDUFA9, known to promote CRC invasion (A).
G alt allele is associated with decreased 5hmC (teal) and increased 5mC (red). Resolving methylation (B) in a 5-Base space masks distinct 5mC and 5hmC trajectories in the promoter flanking region of Cyclin-I, a potential CRC neoantigen.
These distinct modalities are revealed when we resolve (C) in a 6-Base space allowing the detection of otherwise cryptic 5mC and 5hmC transitions.
duet multiomics solution evoC is fully compatible with commercially available Twist hybrid capture panels. Targets are uniformly covered to a high depth without comprising the core performance of duet evoC. Targeted sequencing enhances the already impressive ability of duet evoC to robustly obtain multimodal, phased genetic and epigenetic information, from a single low-input sample of DNA.
duet evoC in 6-Base space detects distinct patterns of 5mC & 5hmC that occur during disease initiation and progression. Targeted duet evoC has the resolution to disentangle the interactions between genetics and epigenetics, detecting Variant Associated Methylation including allele-specific hydroxymethylation.
duet evoC is now available to pre-order as a product from biomodal.
duet evoC also features the following posters if you would like to hear more about its applications: